In this tutorial, you will analyze hyperspectral images of plastic bags with a mixture of powders made from baking soda, vanilla sugar, and potato starch. The tutorial images contain samples with a known amount (%) of these powders, that will be used as the training data set, and samples with unknown powder, where the mixture content will be determined.
Our goal is to learn how to use Breeze to make a quantification model (PLS) and then use it to predict new samples.
|
Steps included in tutorial |
|||||
|
Record |
Model |
Play |
|||
|
|
|
||||
|
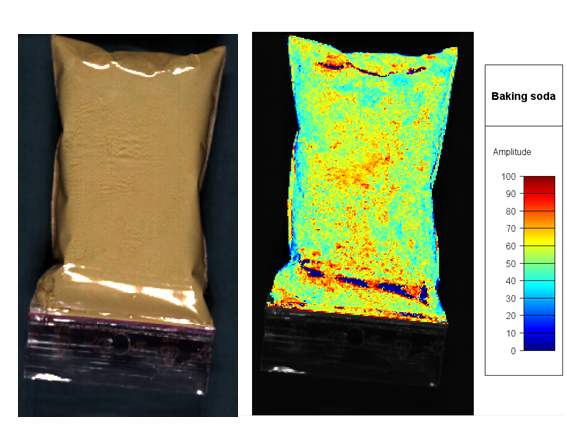
Hyperspectral image SWIR camera
(data was reduced to 67 spectral band to reduce file size) |
|
Start tutorial and download powder measurements
Start Breeze with the shortcut created after installation.
The Breeze start screen should look like this:

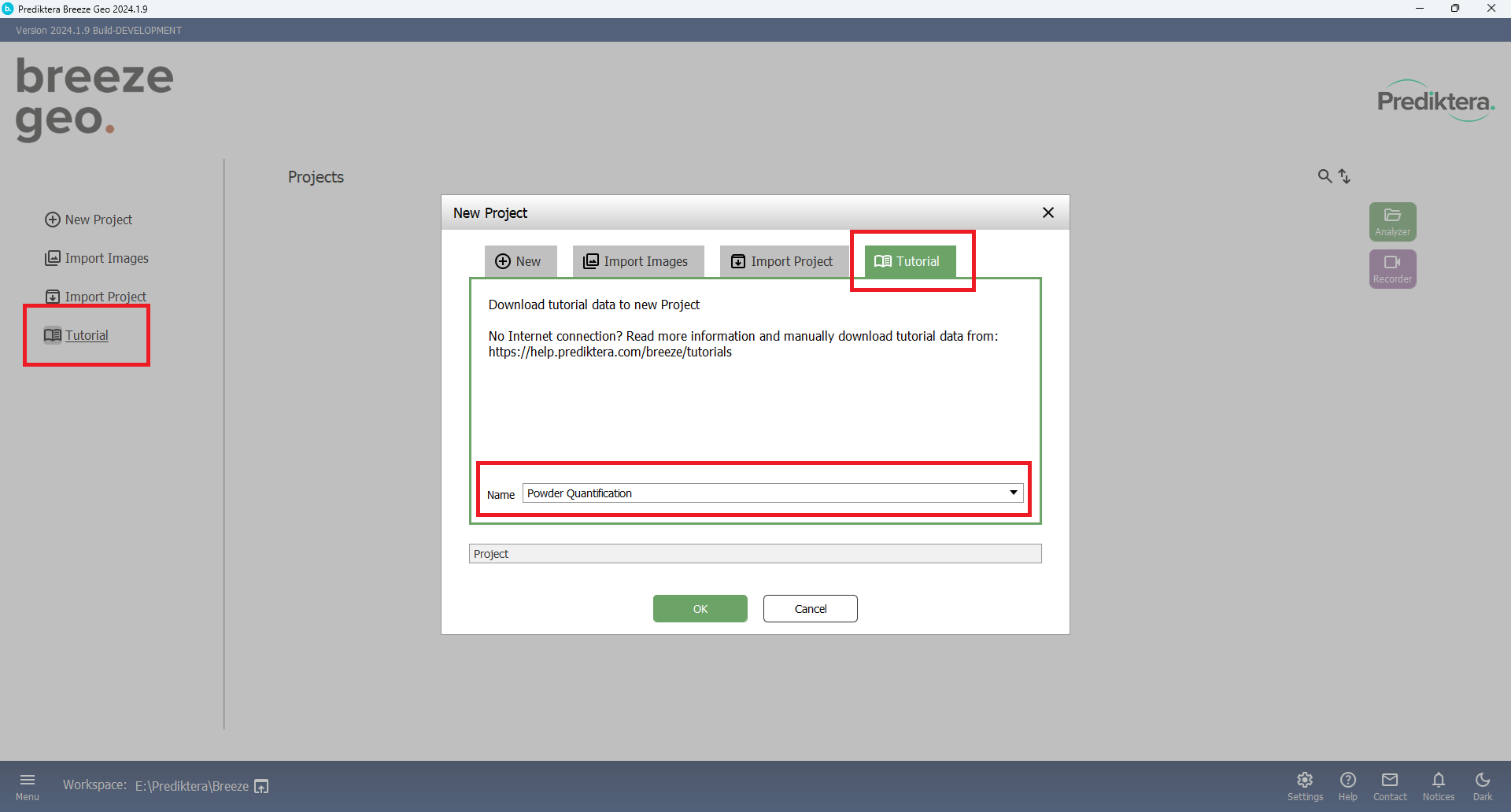
Press the “Tutorial” button and Select the “Tutorial” tab. Select “Powder Quantification” in the “Name” drop-down menu. Press “OK” to start downloading the image data.
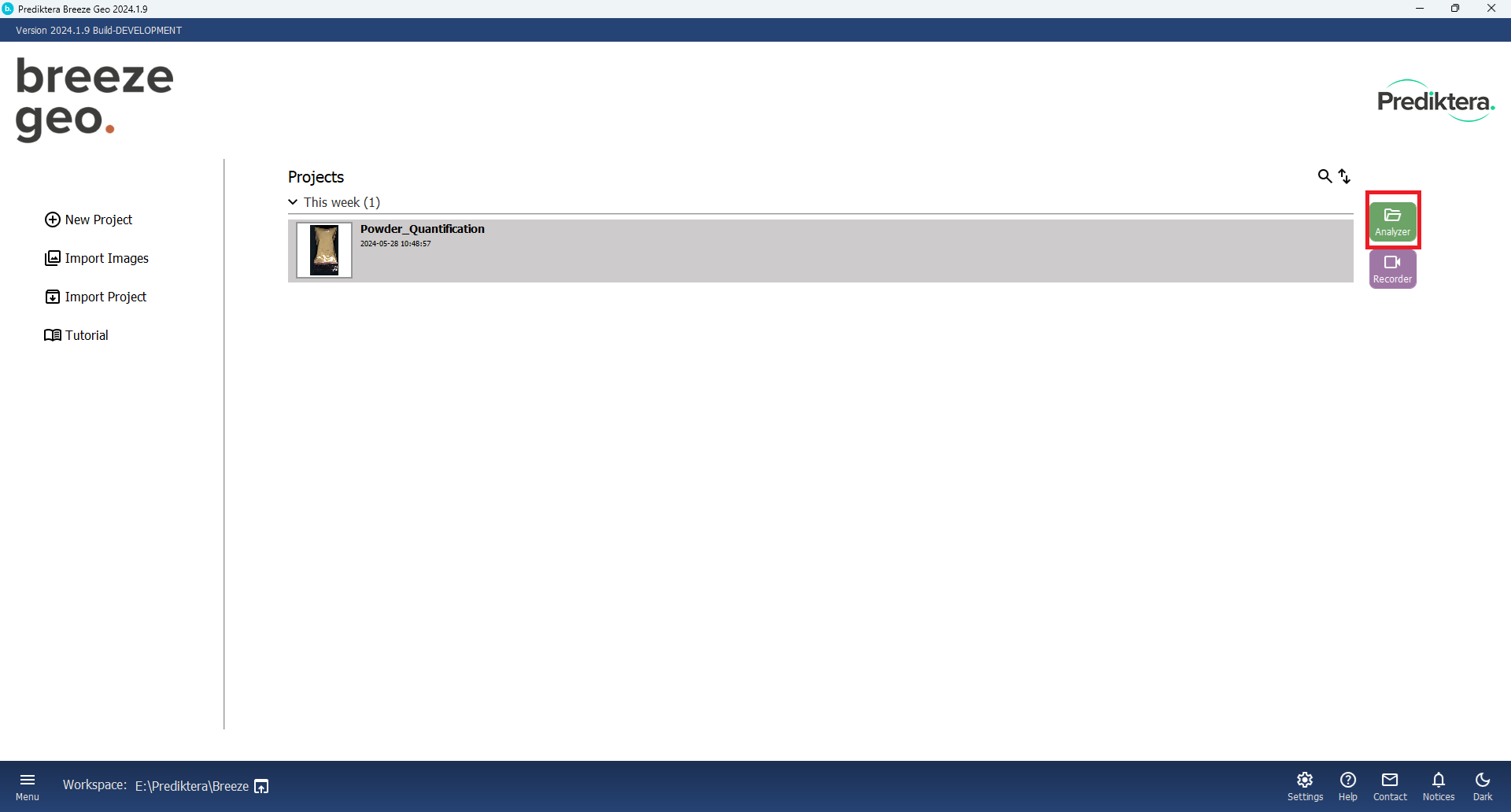
After the Tutorial data is downloaded. Click on “Analyzer” button:
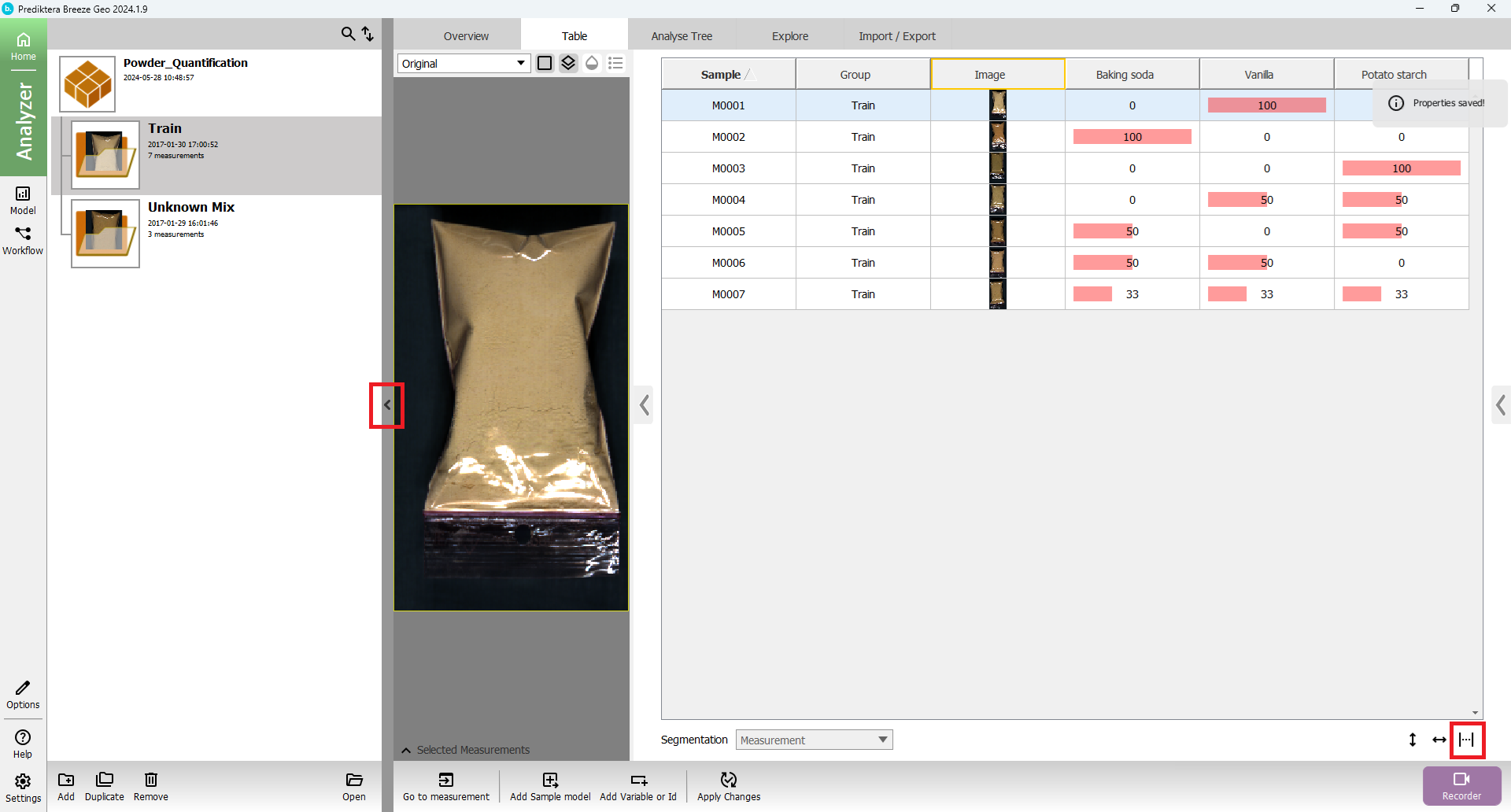
You will see the following table:
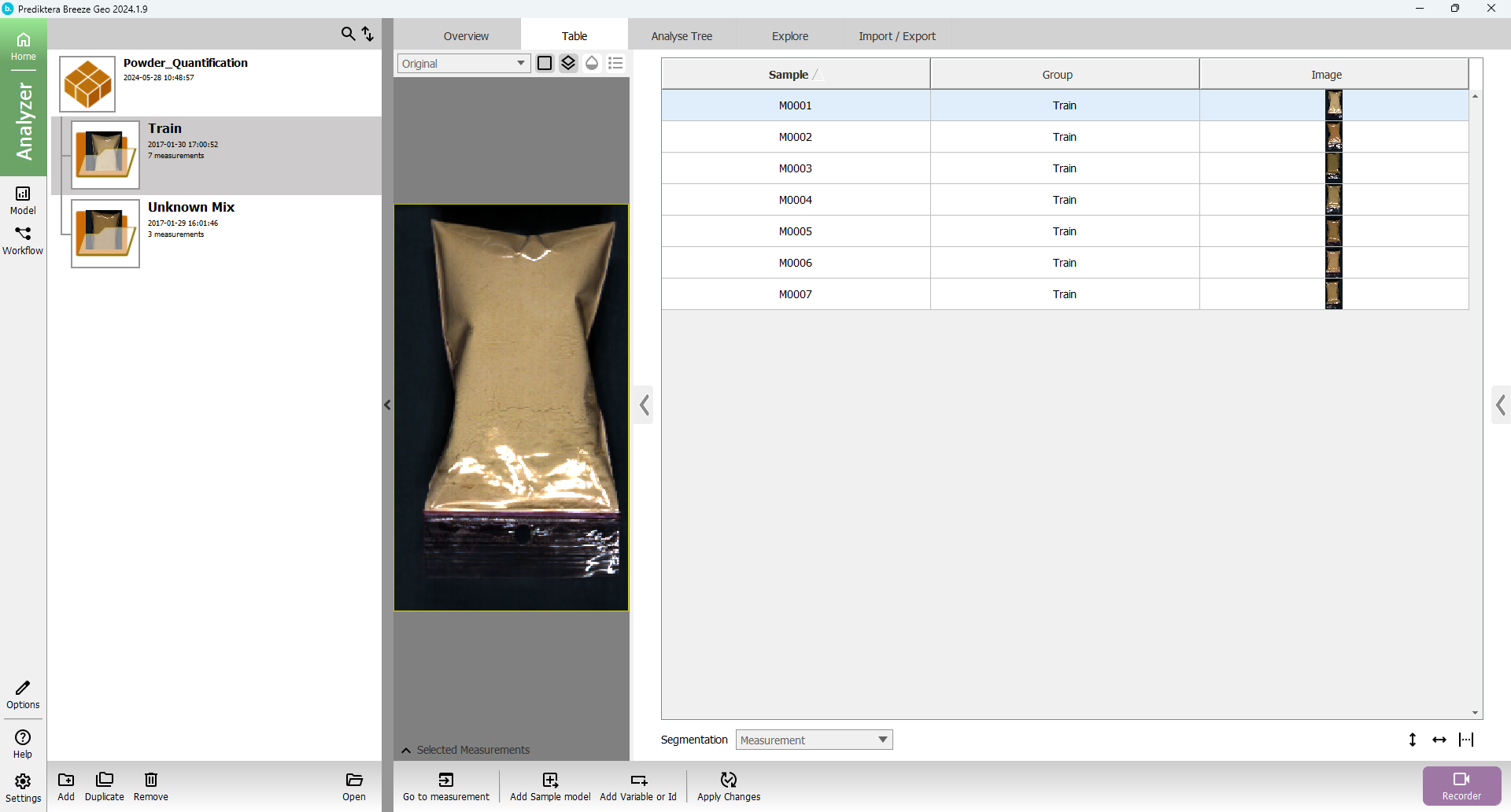
A “Project” called “Powder_Quantification” has now been created that includes seven training images (with different concentrations) and three unknown test images.
The image data in this project is organized into two Groups called Train and Unknown Mix
You can click on a table row to see the preview image (pseudo-RGB) for each image.
Click on “Open” to open the project
Press “Open” button again to open the Train group
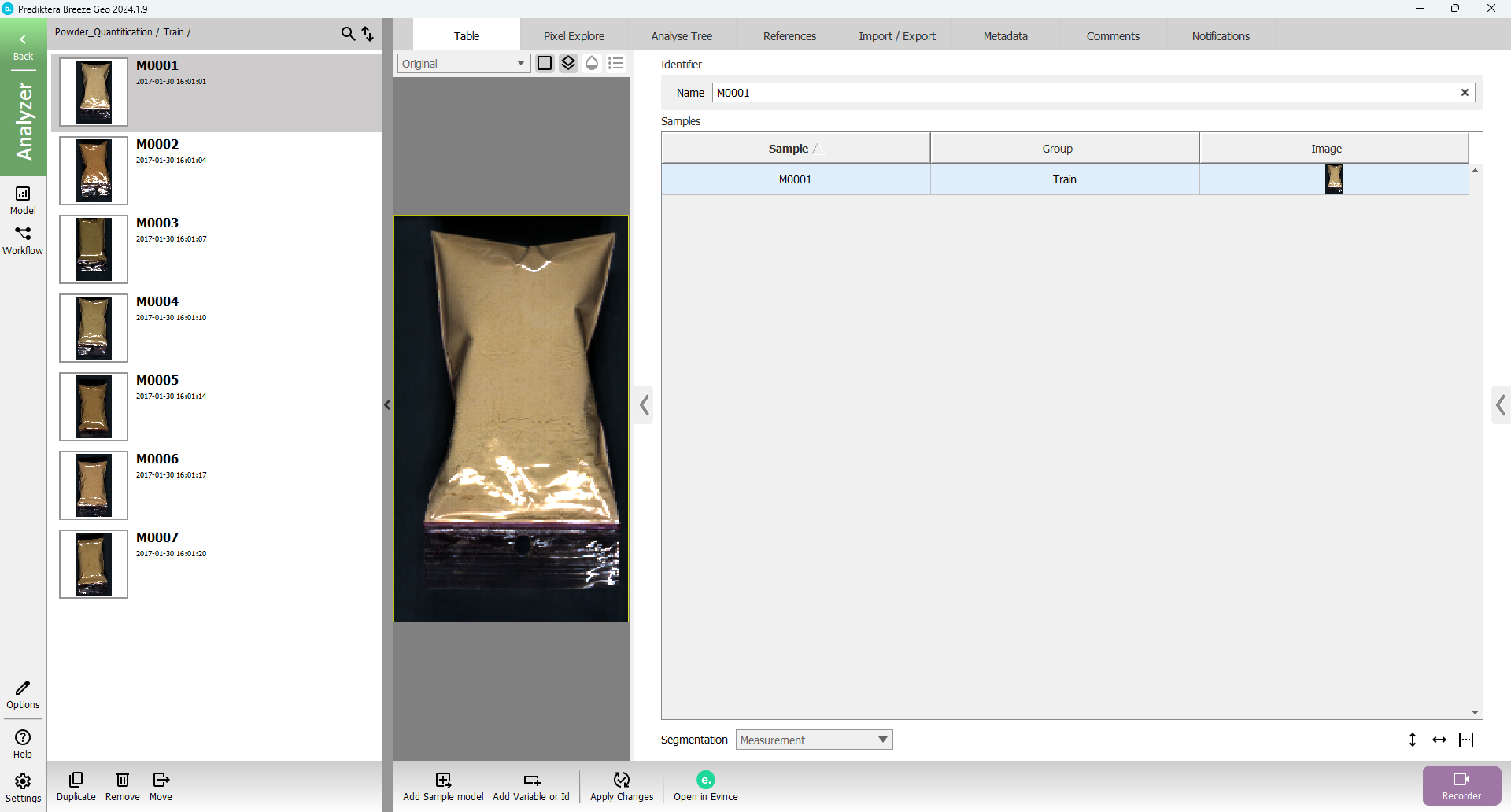
In the menu on the left side, you can now see all the individual images (called “Measurements” in Breeze) in this group.
Click on the “Pixel Explore” tab and click on “Create” button
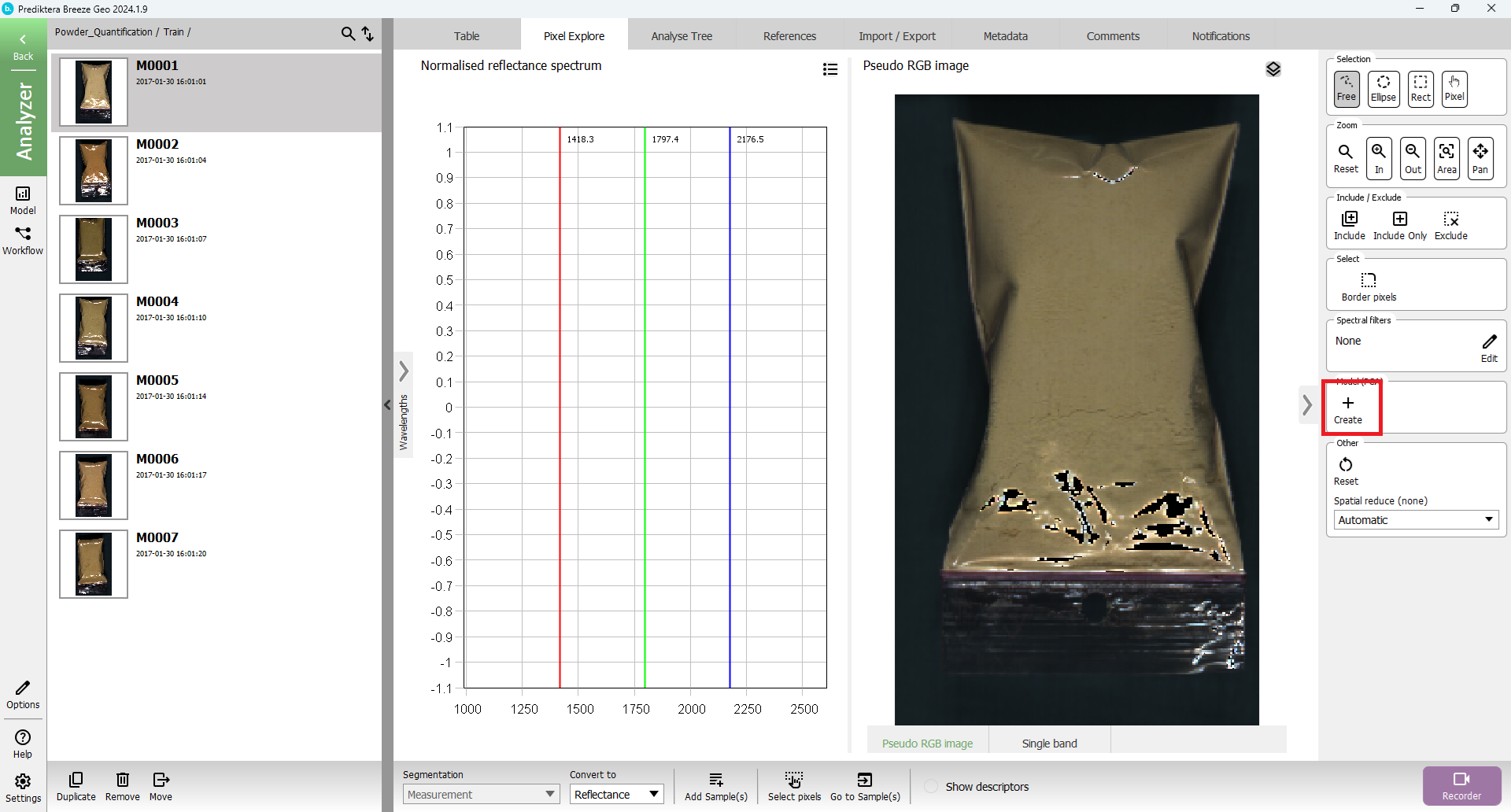
Then you should see this view:
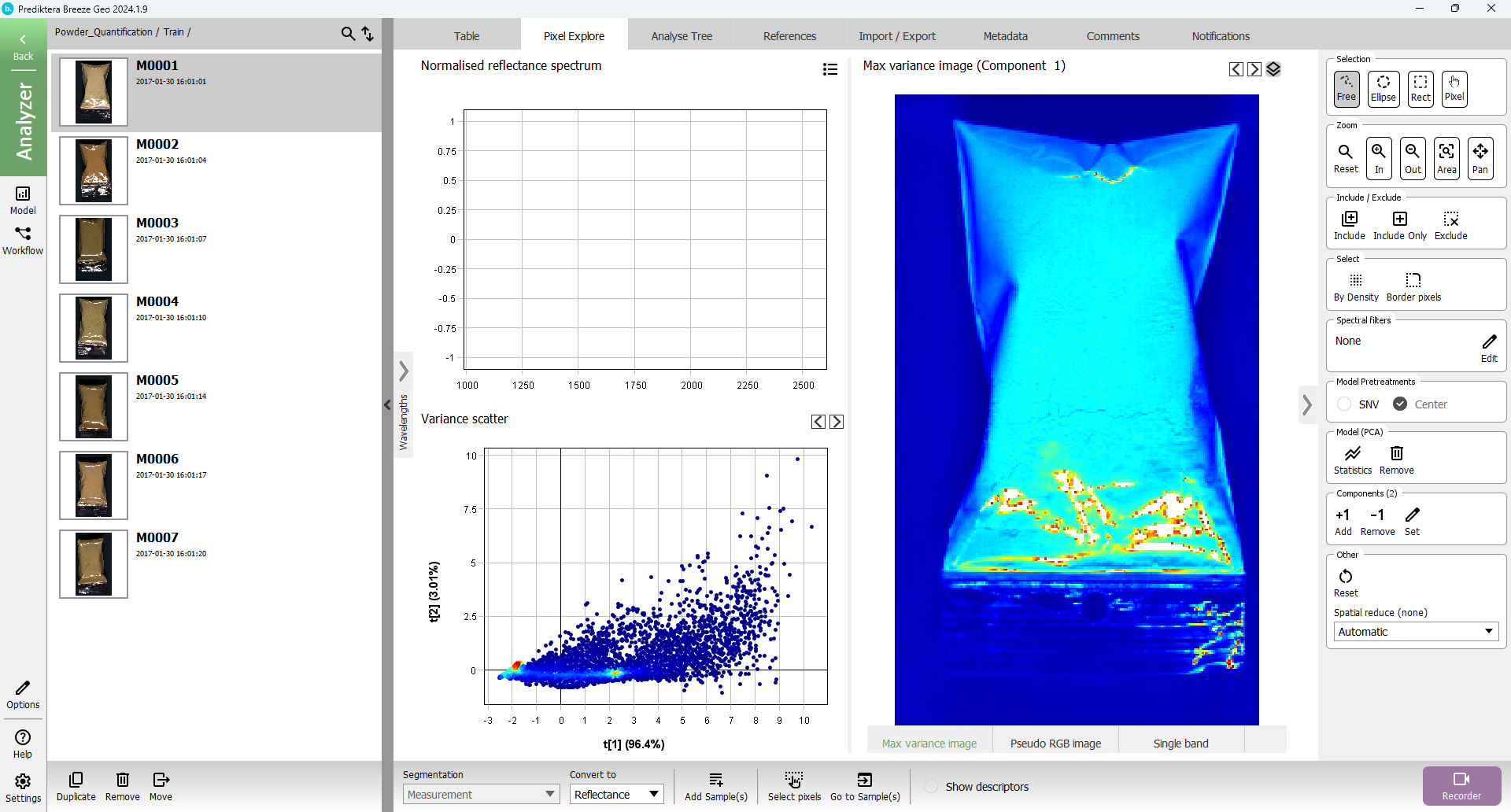
To do a quick analysis of the spectral variation in the image, a PCA model has been created based on all pixels in the image. Each point in the “Variance scatter” plot corresponds to a pixel in the image. The points in the scatter plot are clustered based on spectral similarity. The coloring of the points is showing how closely the points are clustered (red=tightly clustered points).
The “Max variance image” is colored by the variation in the 1st component of the PCA model (the X-axis in the scatter plot, i.e. “t1”), and visualizes the biggest spectral variation in the image. In this case this is the difference between the sample (blue) and the background (red).
Hold down the left mouse button to do a selection of a cluster of points in the “Variance scatter” plot. The corresponding pixels will be highlighted in the image. Move the mouse around in the image to see the spectral profile for individual pixels or use the mouse to do a selection to see the average spectrum for several pixels.
Press the “Back” button in the upper left corner to return to the Group level.

Import of values of powder content for the training set
Press the “Import” tab, select to “Variables and id data” and press “Apply”.
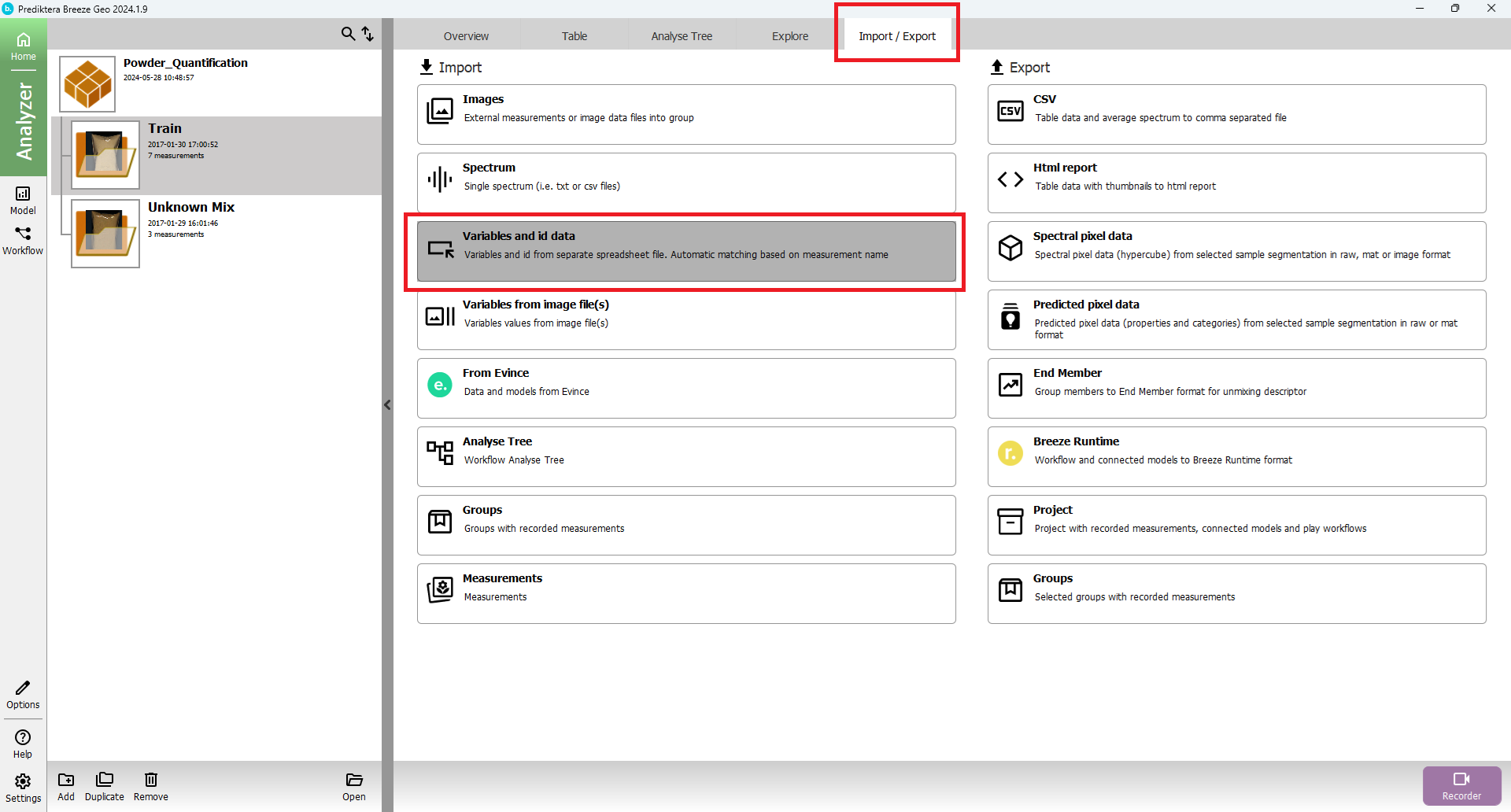
Select “Powder_Quantification_Train.csv” and press “Next”
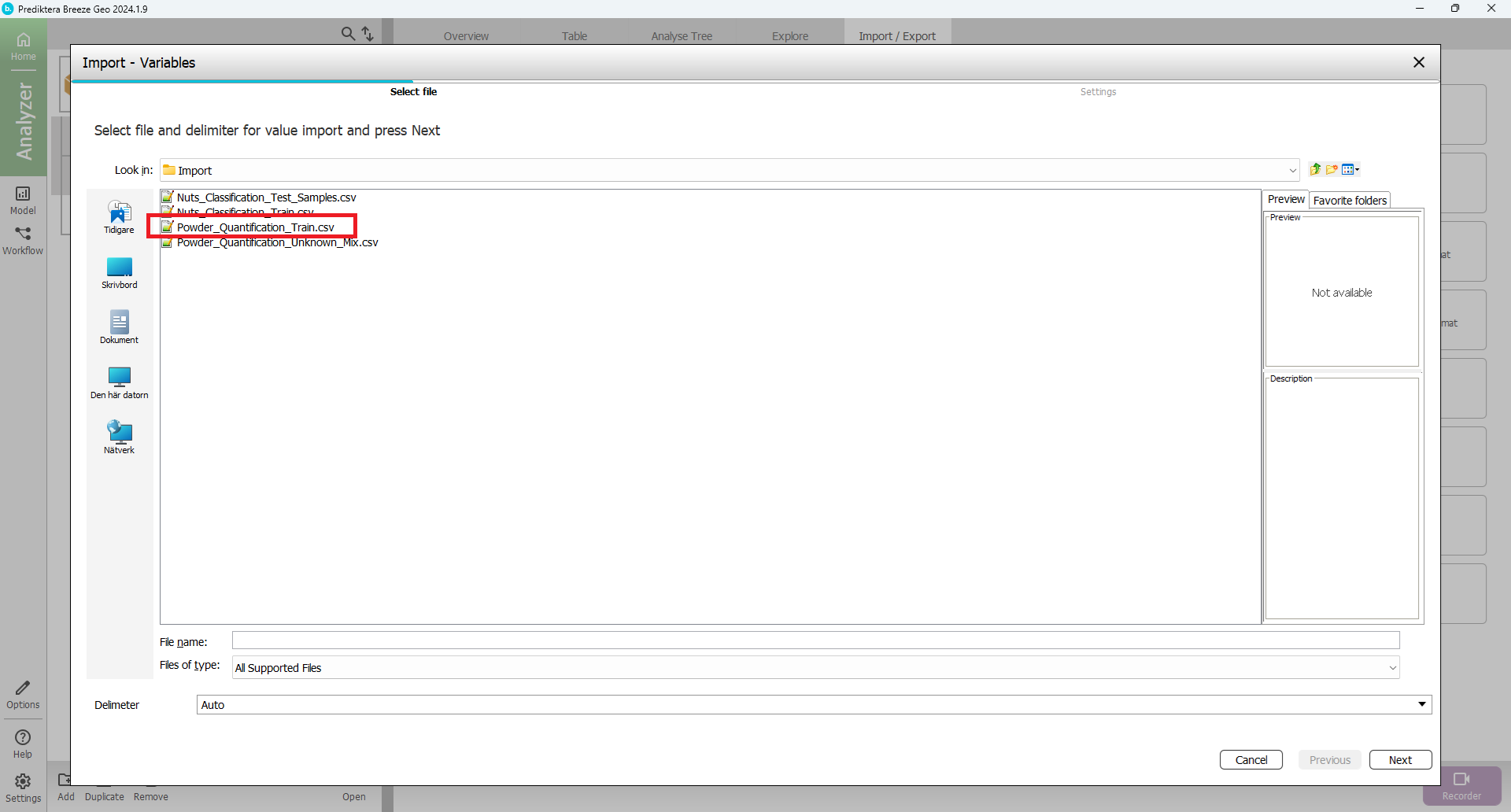
In the next step Press “Finish”
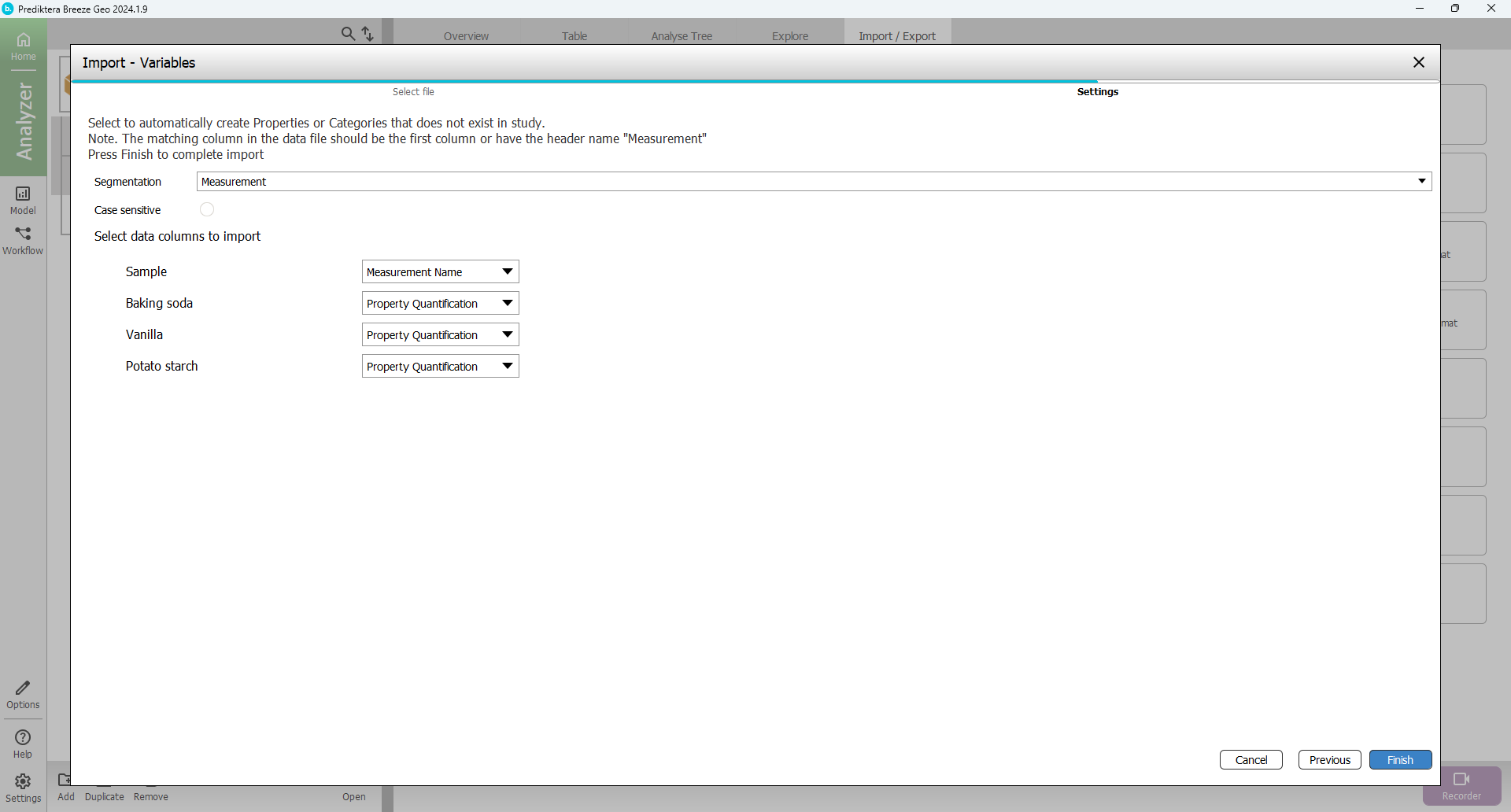
The view should then look like this with columns and values added for the three properties (% of each powder type) for all samples:
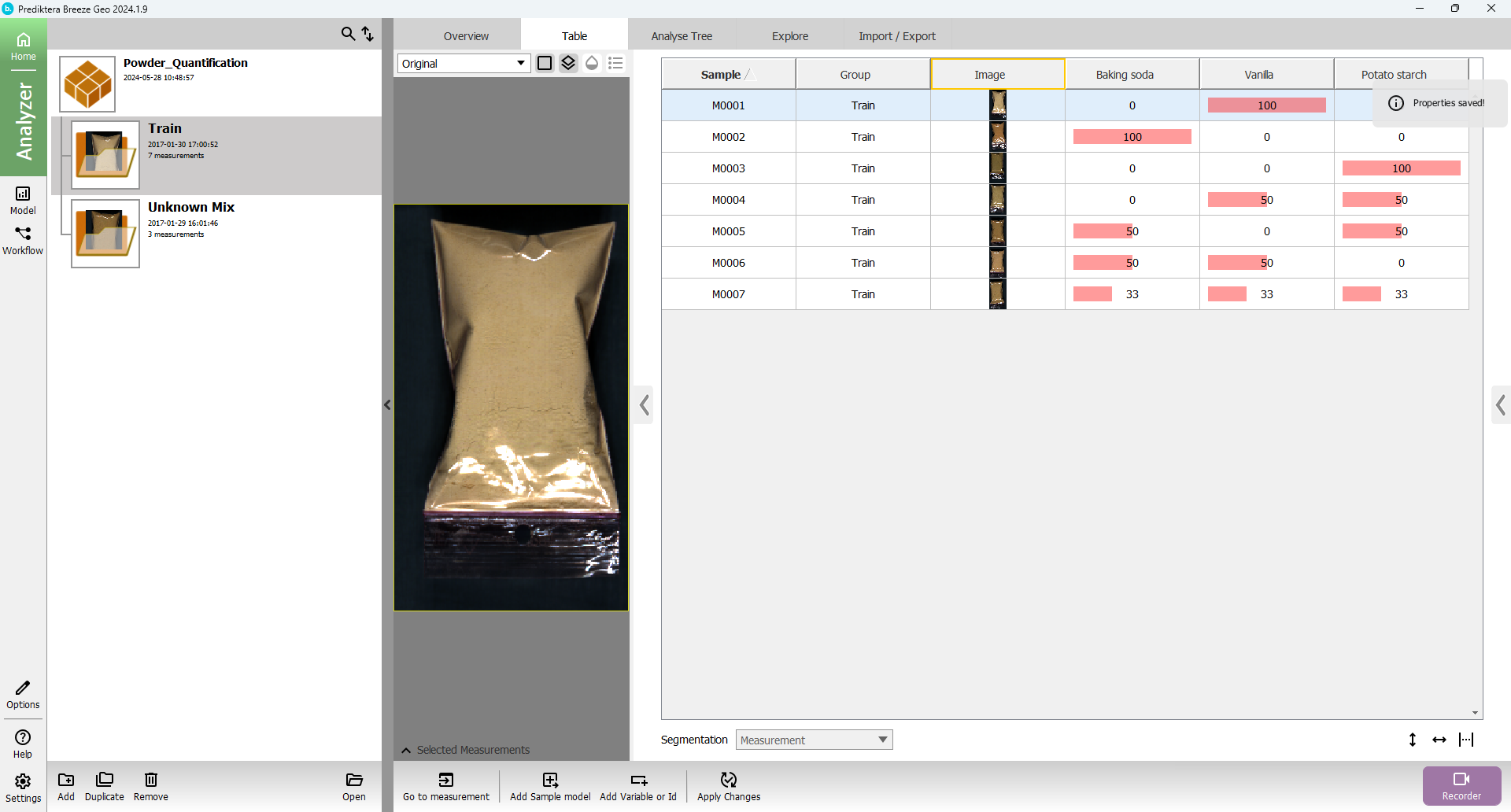
(Hide the Project list by pressing the three dots on the vertical band.
Click on auto adjust table width to show all columns)
If you need to delete variables or IDs, right-click on the header for the column you want to delete and then press the “Delete” option that will appear.
Create a sample model to remove background pixels
You will now create a sample model that will be used to remove the background pixels and to automatically identify the objects (samples) in the images. Under the table Press “Add Sample model” button.
Write a name for the Sample model (or just use the default name) and press “OK”
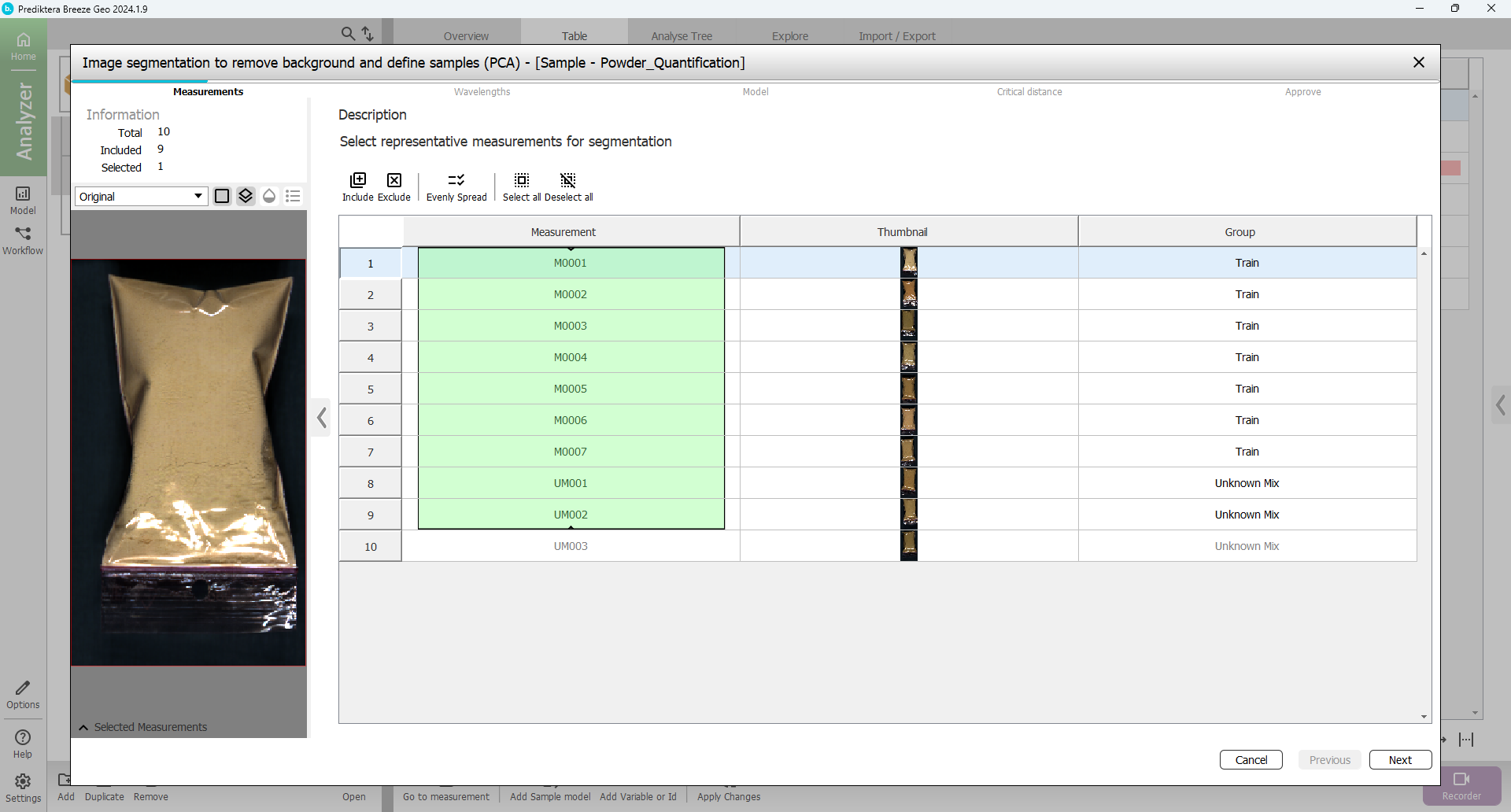
In the first step of the sample model wizard, you can select the images that you will use in the model. By default, 9 measurements are included, which is ok.
Press “Next”
In the next step of the wizard, you can select spectral bands (wavelengths) to use in the model. By default, all wavelengths are included and SNV is used as the pretreatment.
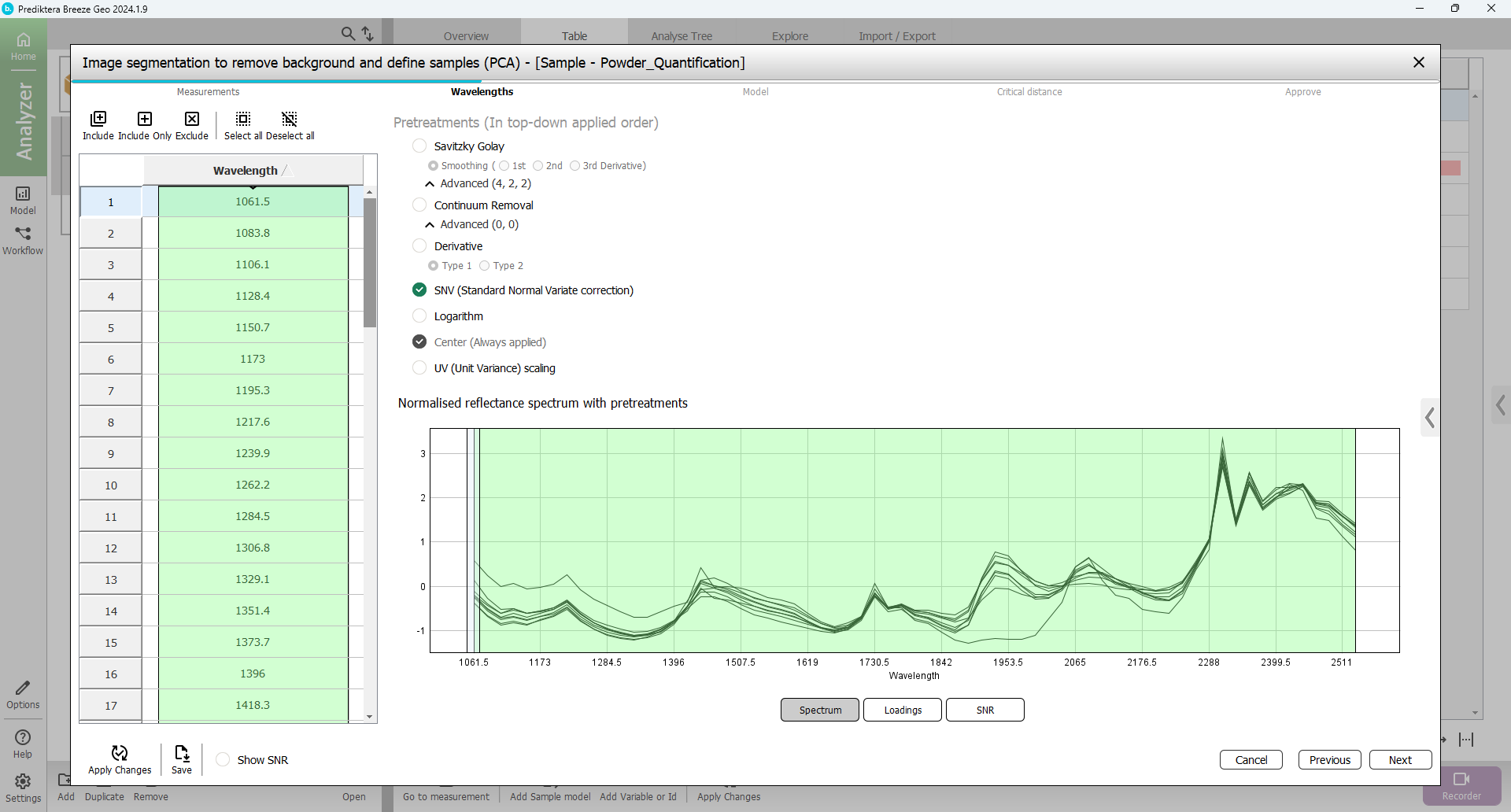
You can play around with different pretreatment choices and see how the spectrum changes after you
Press “Apply changes”
The following picture is the same spectrum as the pretreatment first derivative from Savitzky Golay.
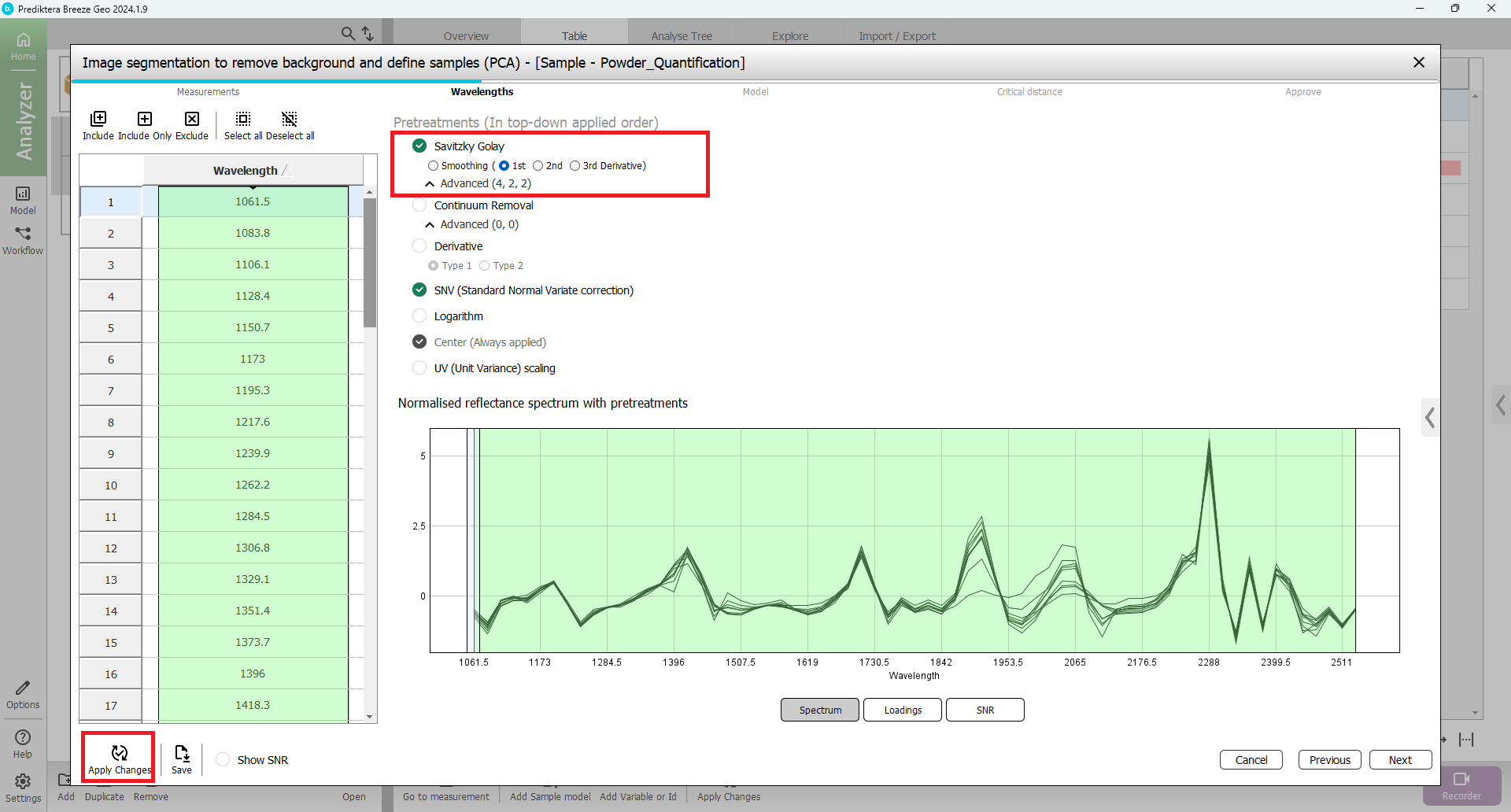
When you are done experimenting with the different pretreatments change back to only including SNV and press “Apply Changes” before you Press “Next”
In the next step, you will select the pixels to use in the sample model.
A mosaic has been created of all images and a PCA model has been created from all pixels in this mosaic. Select a region containing only bag sample pixels by holding down the left mouse button and marking the area inside one of the samples. You can zoom in by using the mouse scroll wheel (or the Zoom tools that be selected in the menu on the right.)
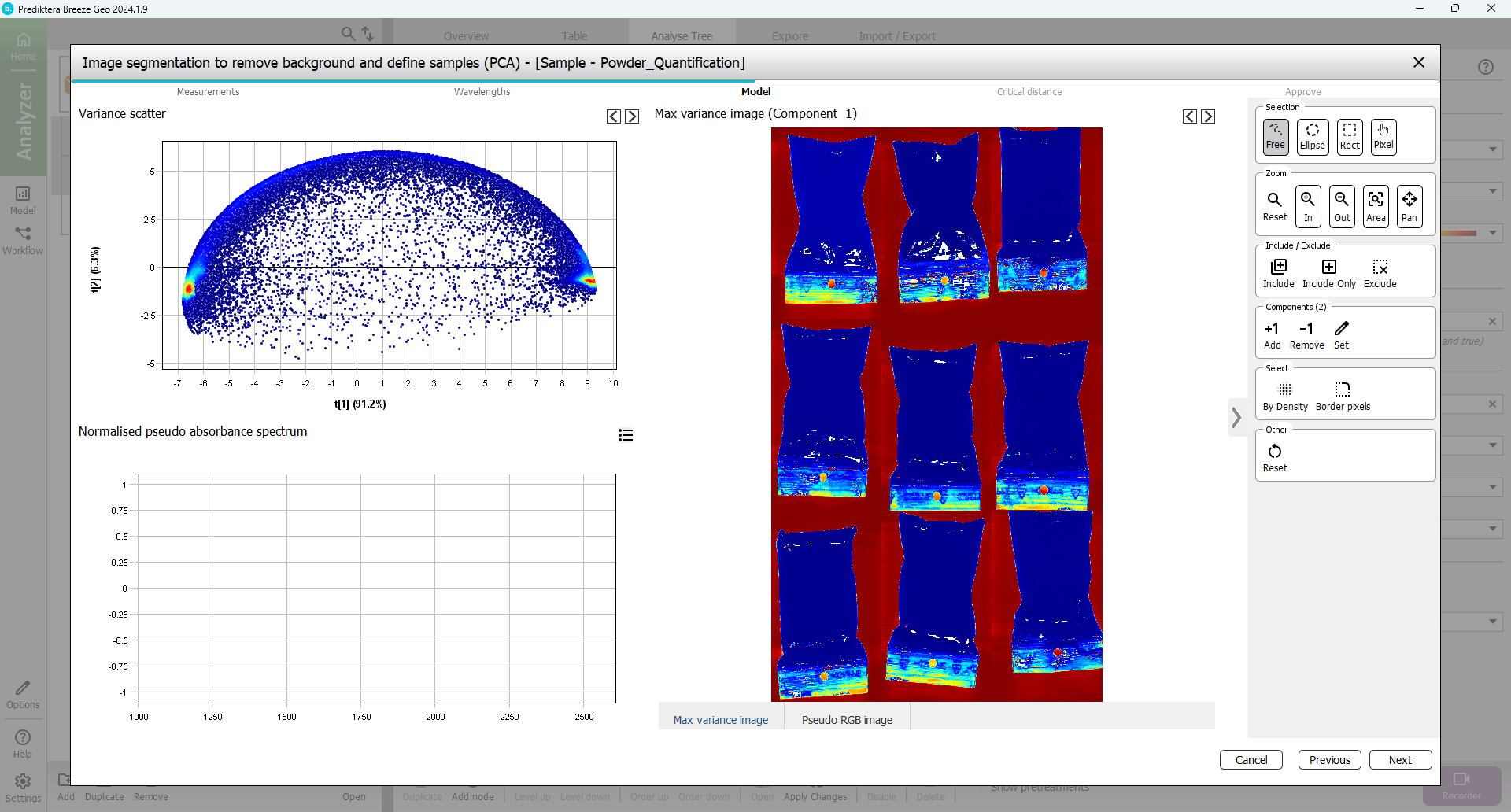
The corresponding pixels are selected on the Scatter plot to the left.
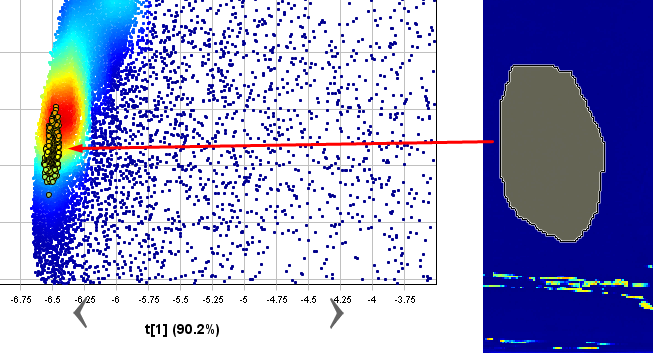
Now you know that the powder pixels are in the cluster on the left side of the Scatter plot. In the scatter plot, select all points in the cluster on the left side (use the pixel density coloring red, yellow, green and light blue as help).
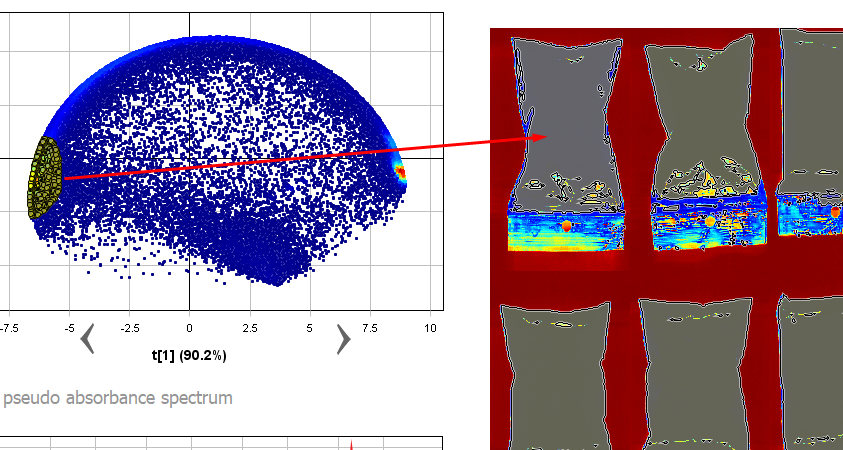
Now you can see that the selected pixels are highlighted in the image, and evidently these pixels belong to the samples.
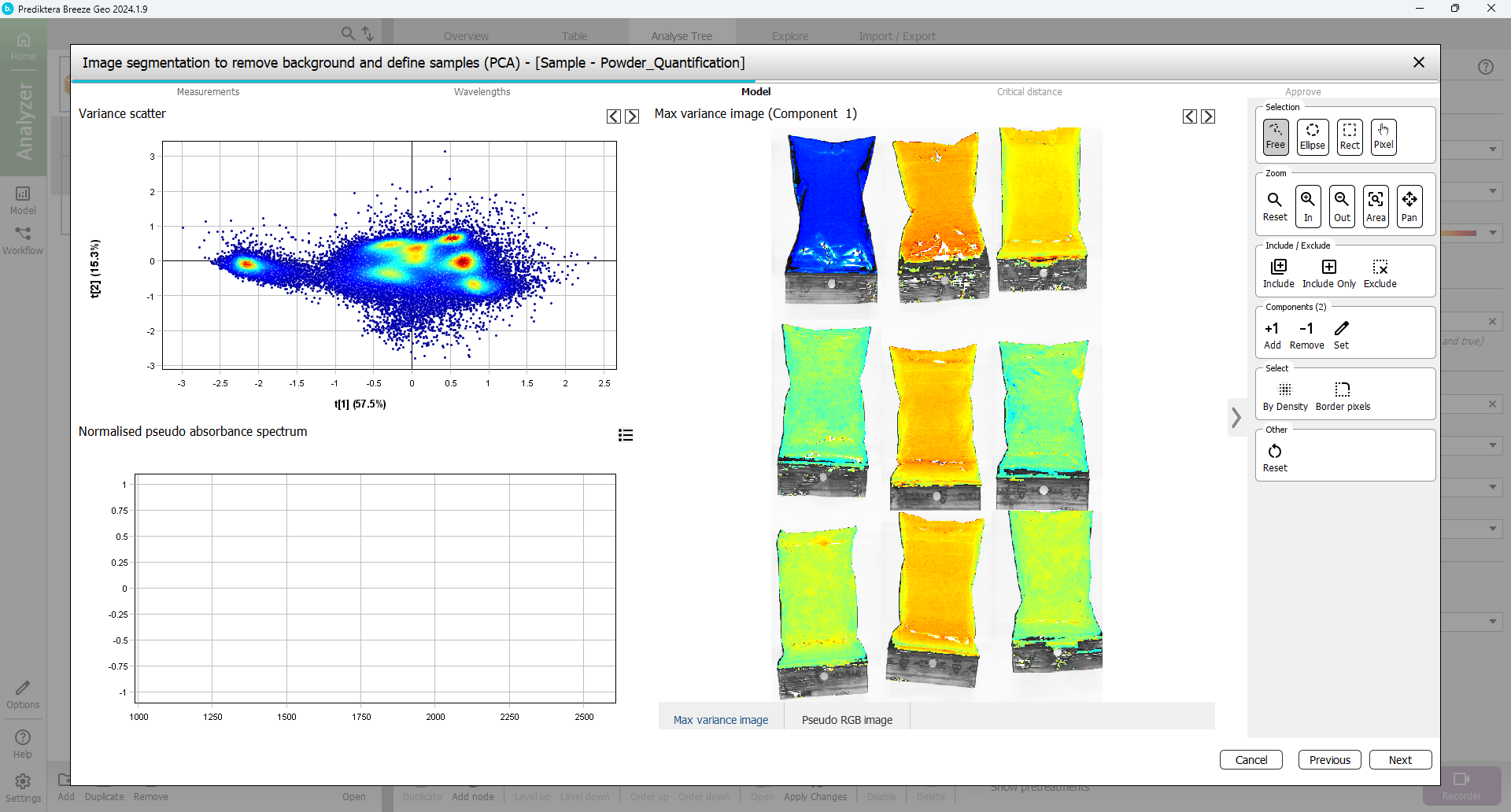
To include only these pixels in the model and exclude all other pixels,
Press “Include only” and wait for the model to be updated.
The plots are now updated and will contain mostly the powder sample pixels.
To clean up the sample pixels even more you can remove the pixels bordering around each sample object. Press the “Select: Border pixels” button.
Use the default of 1 border pixel and press “OK”. The border pixels have now been selected.
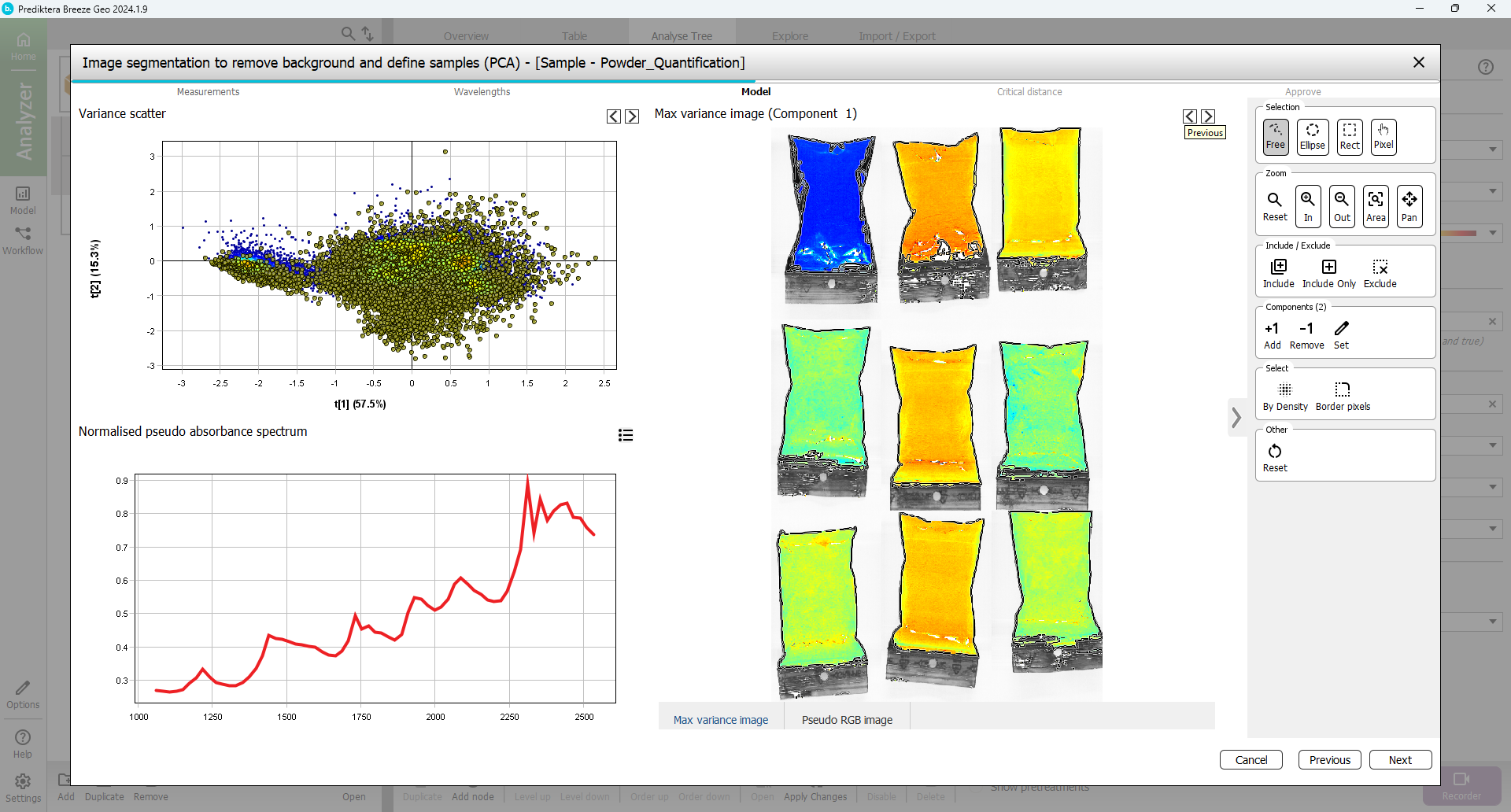
Exclude the border pixels by pressing the “Exclude” button
Press “Next”
In the next step, you will set the Critical Distance threshold. This is the distance to the sample model and will be used to determine if pixels are sample or not. The histogram is showing the distance to the model for all pixels in the images. Pixels on the left side of the red vertical line (critical distance) are inside the threshold.
Drag the red line to the right to move the threshold. As you can see from the image, more pixels are included when doing this. The aim is to find a level where all powder pixels are included but not pixels from the background (As a general recommendation you can use the default threshold shown with a black vertical line).
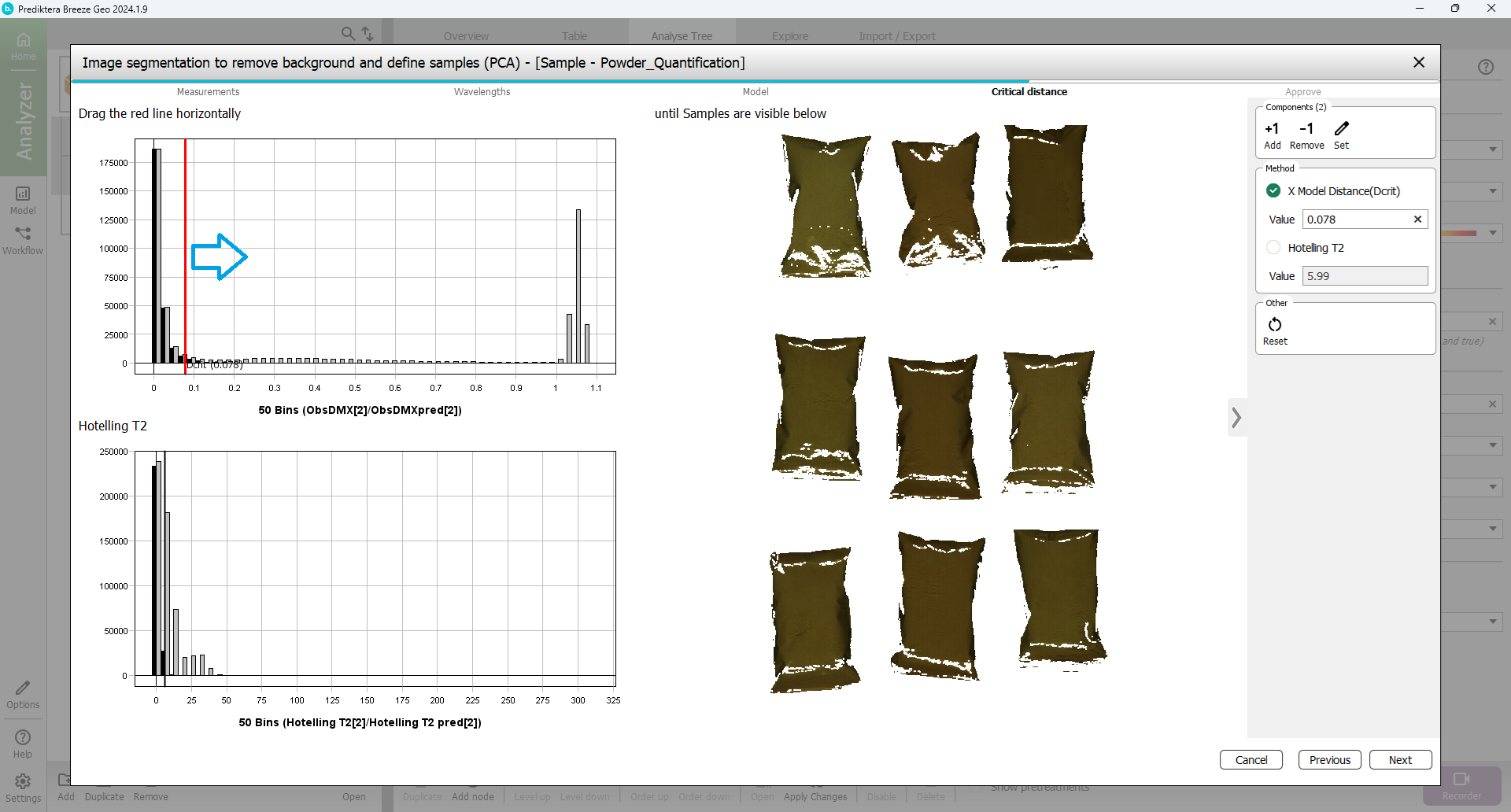
Press “Next”
You are now at the last step of the sample model wizard. The “Minimum area size” is used to automatically exclude smaller unwanted objects (for example dust or dirt). Breeze calculates a suggested minimum area size for your data. In this example, any objects under 2000 pixels will be excluded from the image (depending on how you did the pixel selection this value might vary). A value around 1000-3000 should be OK.
Press “Finish” to create the sample model and apply this to all images in the project.
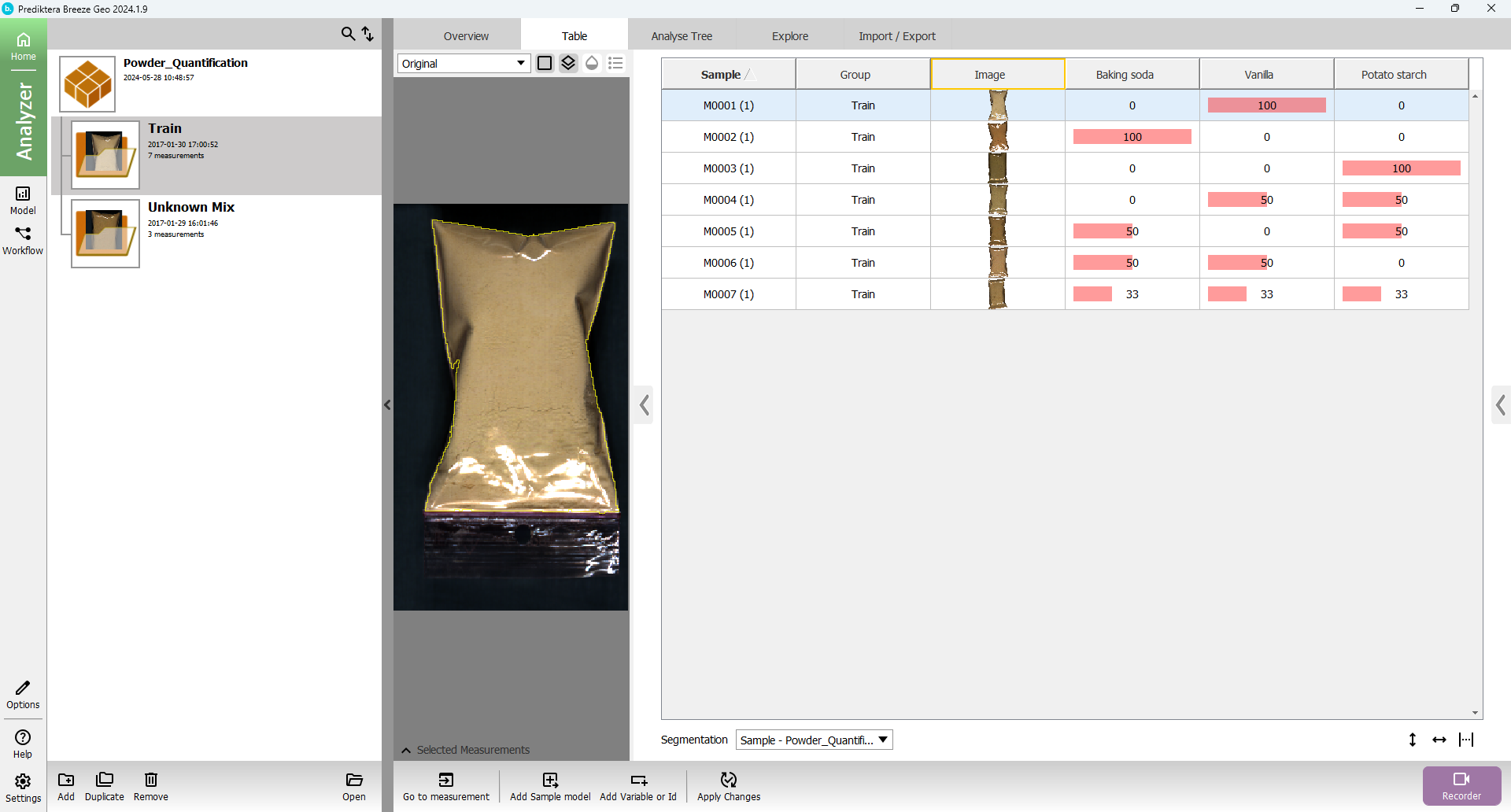
In “Table” for the project, you can now see all the sample objects in the images after the sample model has been applied and the background pixels removed.
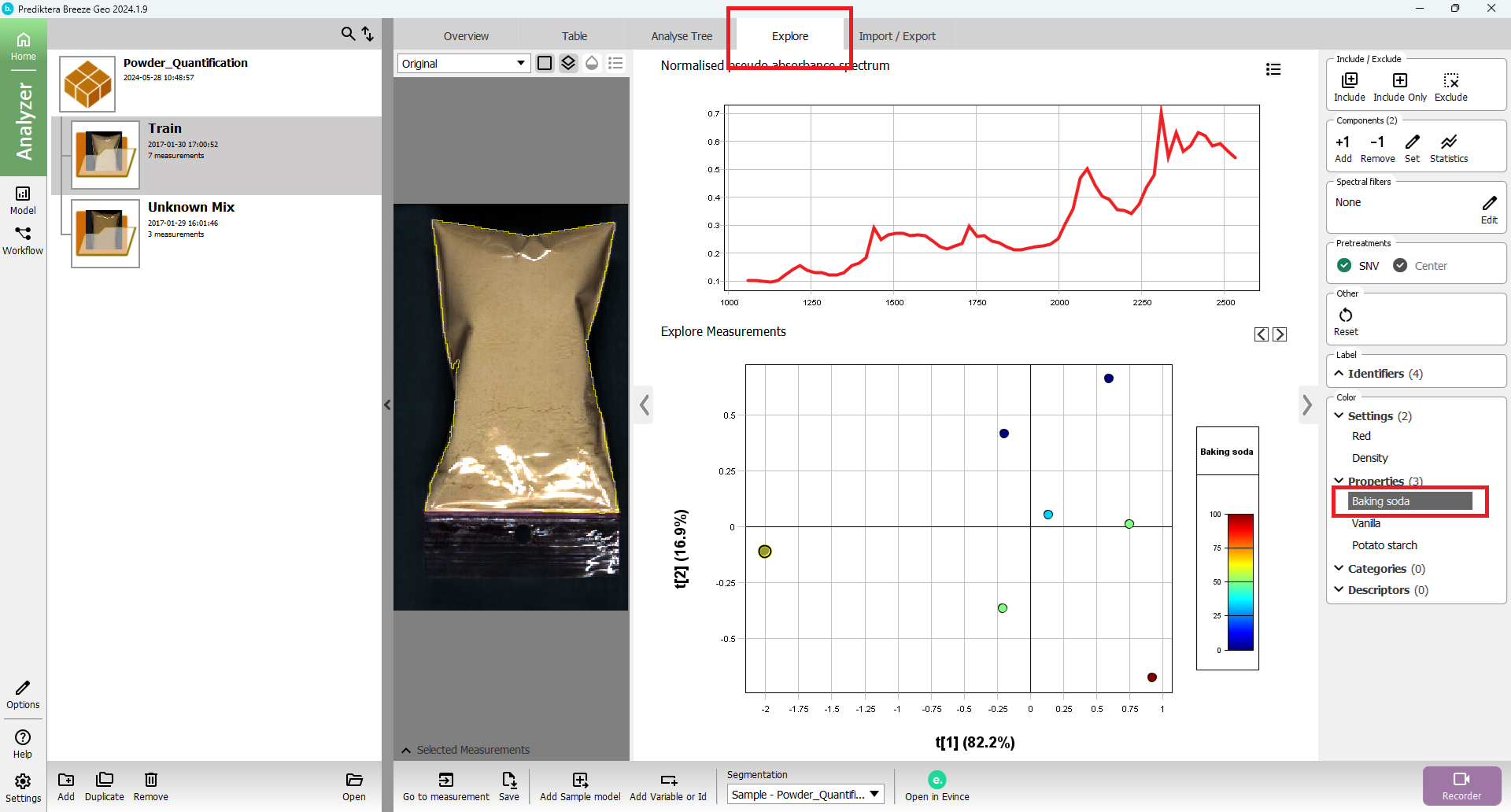
Press the “Explore” tab. A PCA model has been created based on the average spectrum for each sample. Each point in the scatter plot corresponds to a sample and the points are clustered based on spectral similarity. Select one or several points to see their average spectrum.
In the menu on the right click on the property name “Baking soda” to color the scatter plot based on the different property values.
Create a PLS quantification model
You will now use the average spectrum for each sample and the property values that you have imported (% of the three powders) to train a quantification model.
Press the “Model” button in the lower right corner of the screen to move to the Model step.

In the menu on the left you can see the Sample model that you created before. To make an additional model Press the “Add” button.
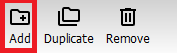
In the window that appears, press the “Quantification” tab.
Write a name for the model (or just use the default name) and press “OK”.
In the first step of the Quantification wizard, you can select the Properties (continuous Y-variables) that you will use to build the model. You can either model one Y variable at a time or include all Y in the same model. The results can vary depending on the data set. In this example, we will model all Y in the same model. Make sure that all three properties are selected and Press “Next”.
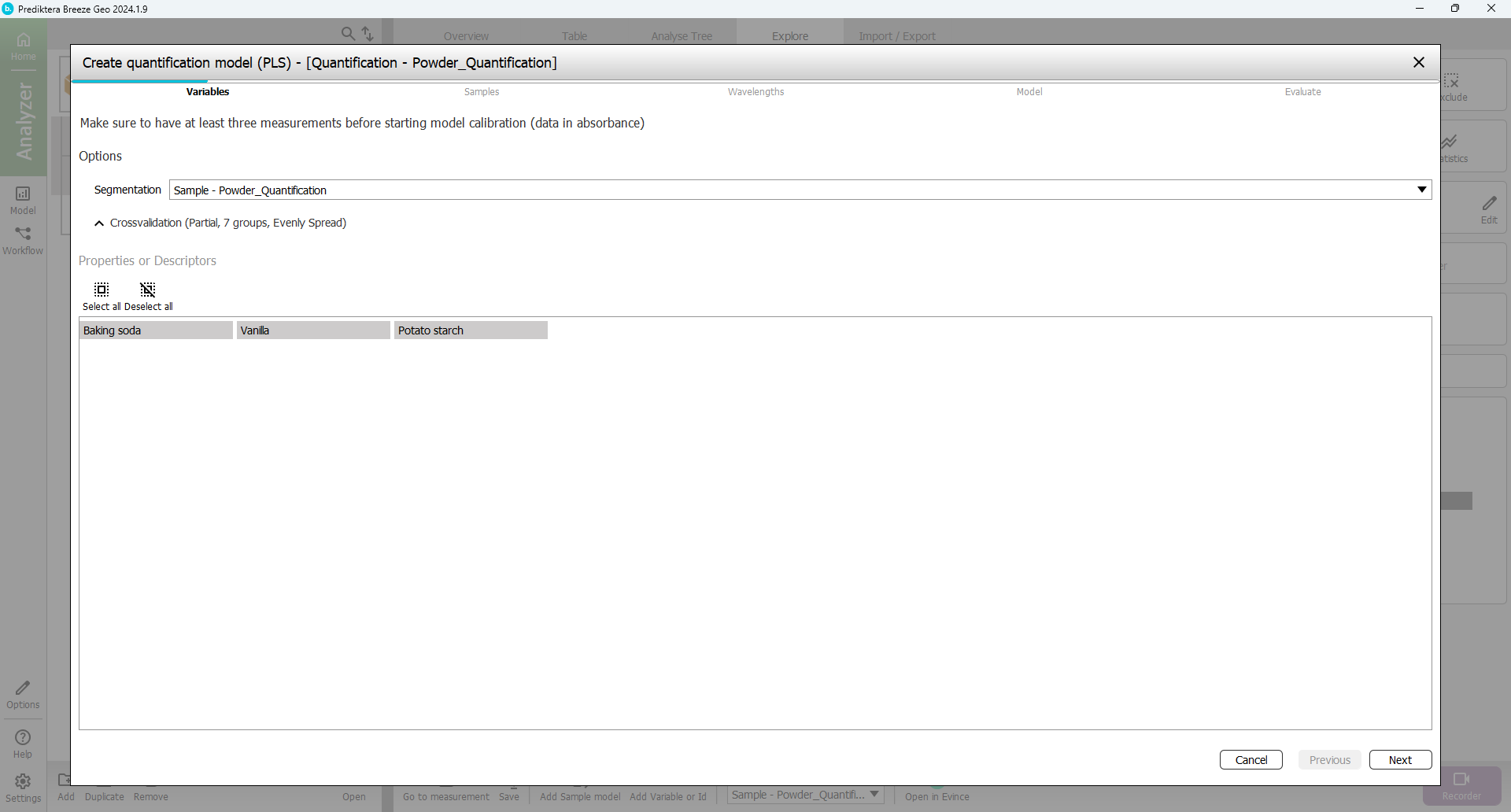
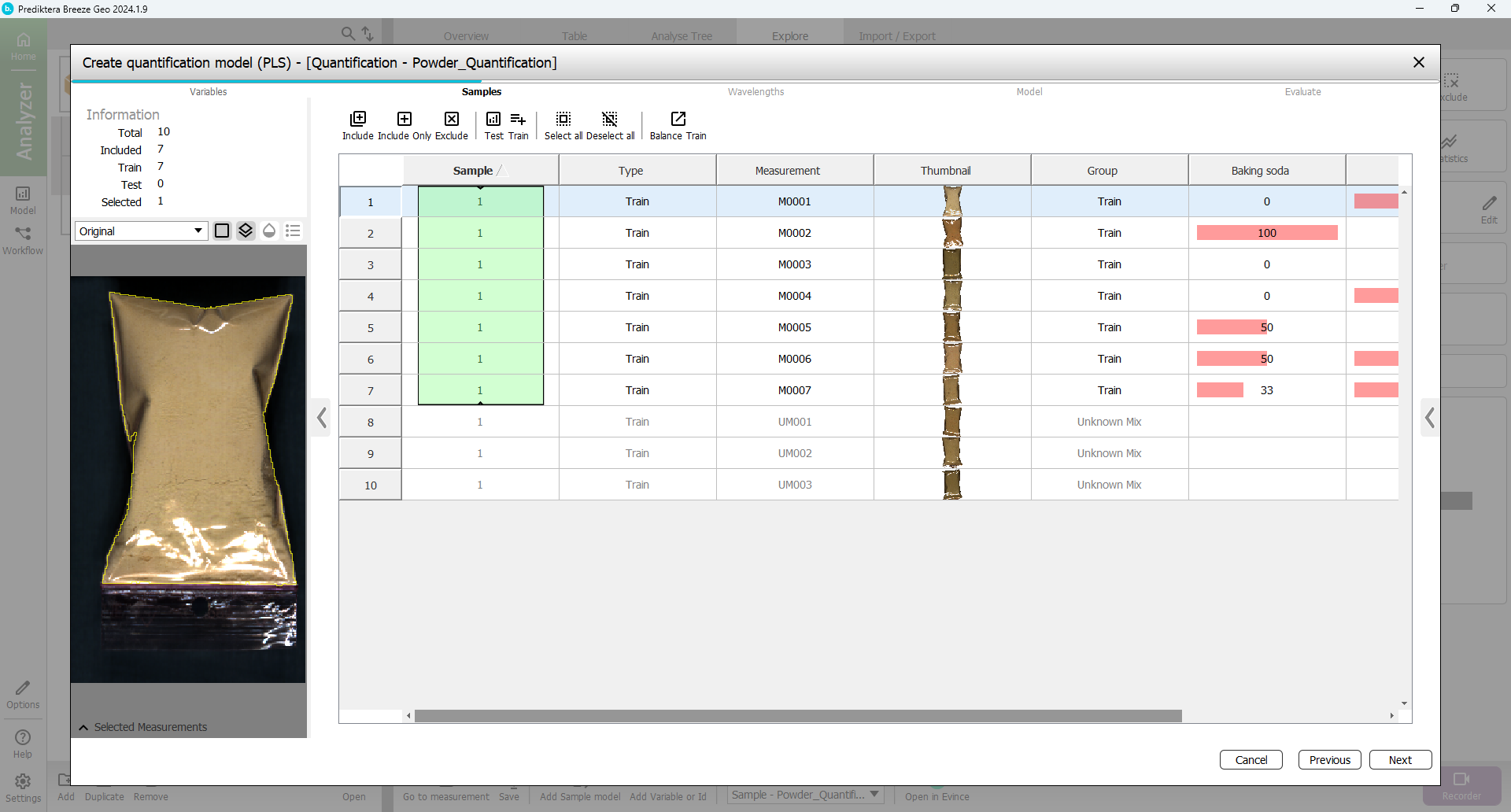
In the next step of the wizard, you can select the samples that you want to include in the model. By default, the “Train” group has been included and the “Unknown Mix” group has been excluded (since these samples do not have any entered data for % of powder they can not be used for the training). This is OK.
Press “Next”.
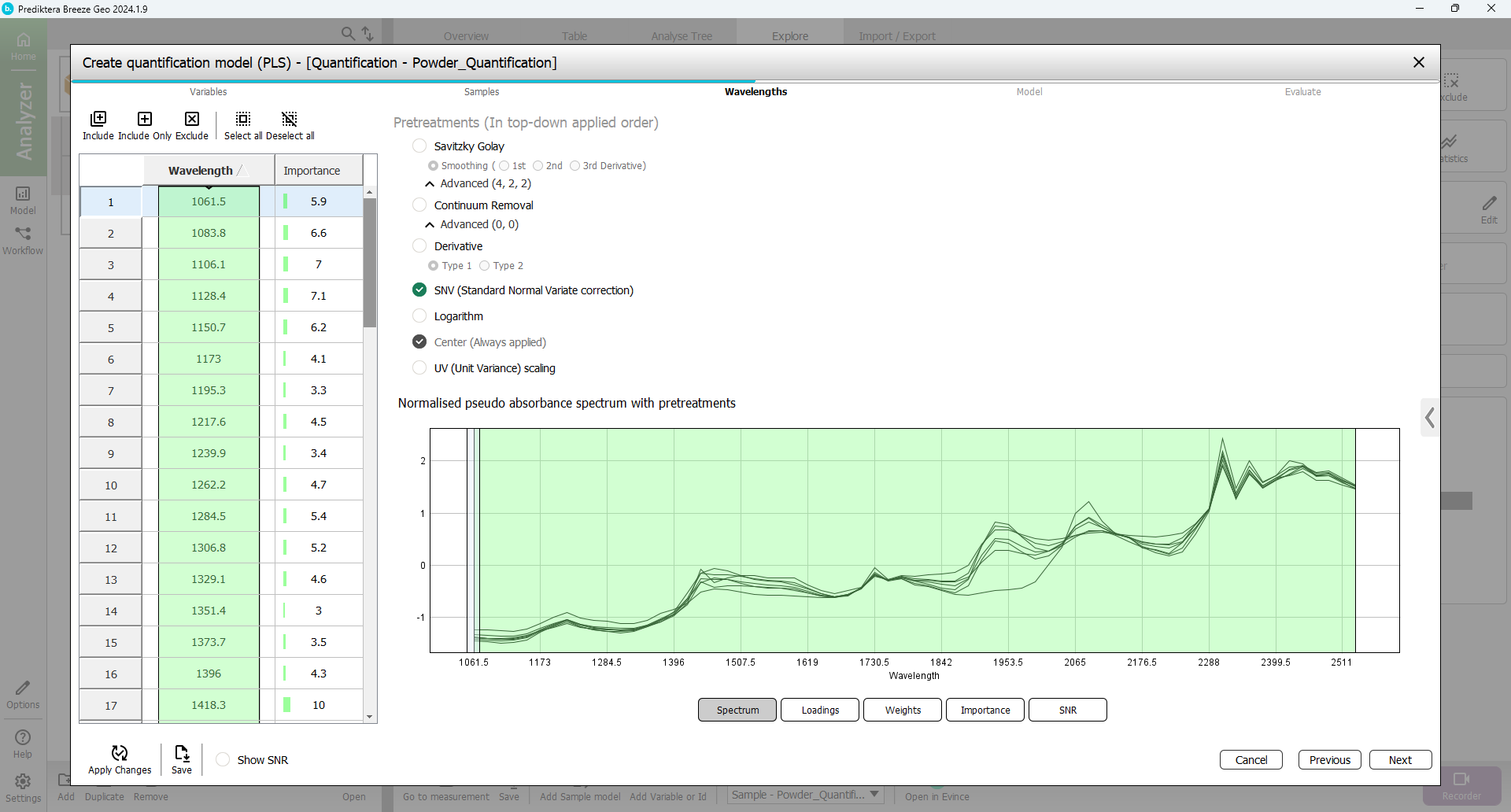
By default, all wavelength bands are included. The graph on the right is showing the average spectrum for each sample. Above this graph, there is an option to select different pretreatments of the spectral data. By default “SNV” is used. All default settings here are OK.
Press “Next”.
Read more on Pretreatments
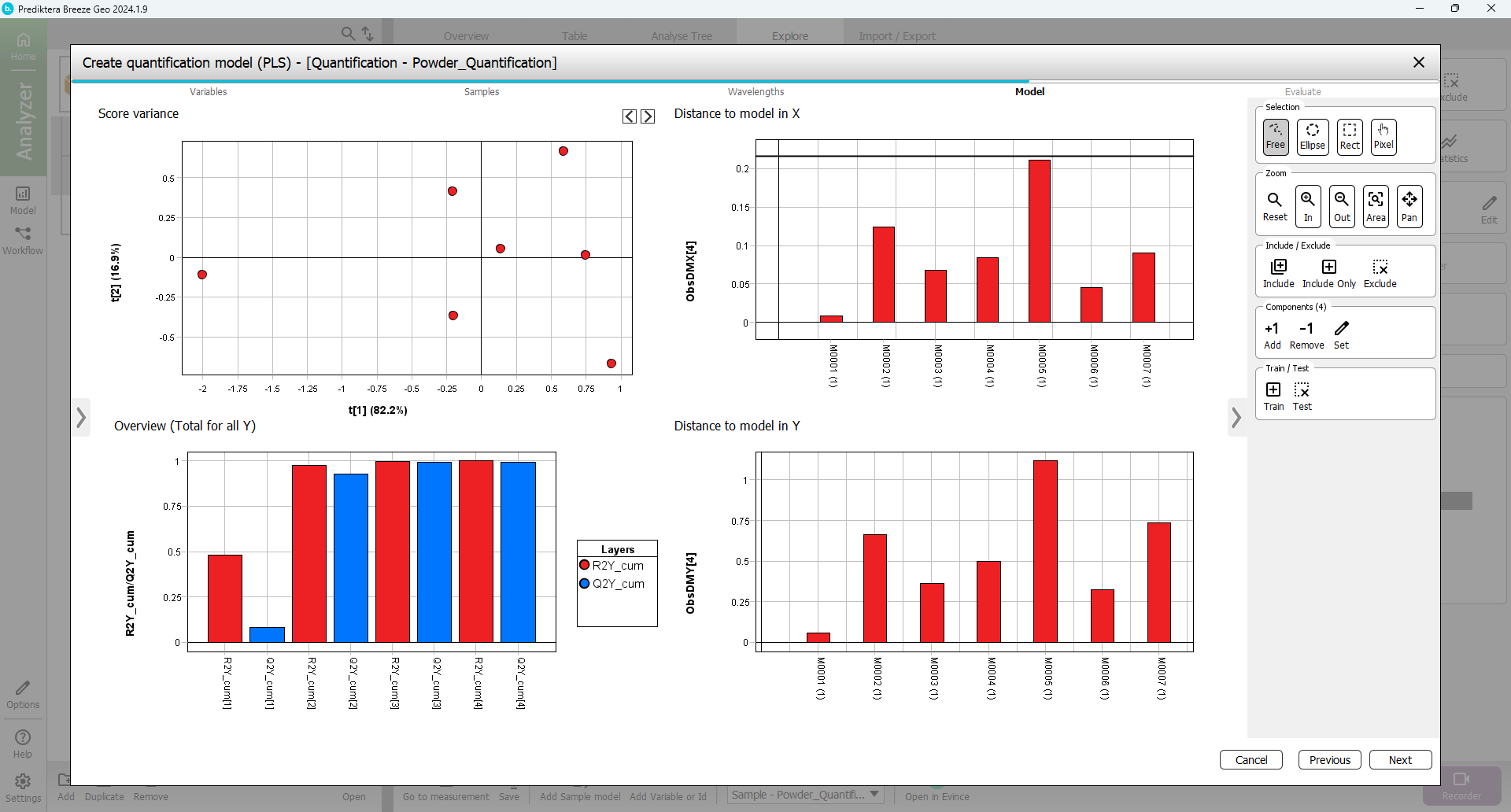
A PLS quantification model has now been calculated.
The “Overview (Total for all Y)” graph is showing how good the PLS model is. It also shows the number of components that were used for the model. In this case, the autofit used four components. The R2 (model fit) and Q2 (prediction from cross-validation) using four components are around 0.99 indicating a very good model. An R2 and Q2 value of 1.0 indicates a perfect model explaining all the variations. A value of 0 indicates that no variation can be explained. (In this case 3 components would be enough but for simplicity we will use the autofit of 4 components.)
'The “Distance to model in X” and “Y” graphs show the distance to the model for each sample. A high bar indicates that the sample might be an outlier (for the X distance use the horizontal black line as a guide. The “Model” scatter plot and the “Distance to model” graphs can be used to identify and exclude outliers.
Since everything looks OK press “Next”.
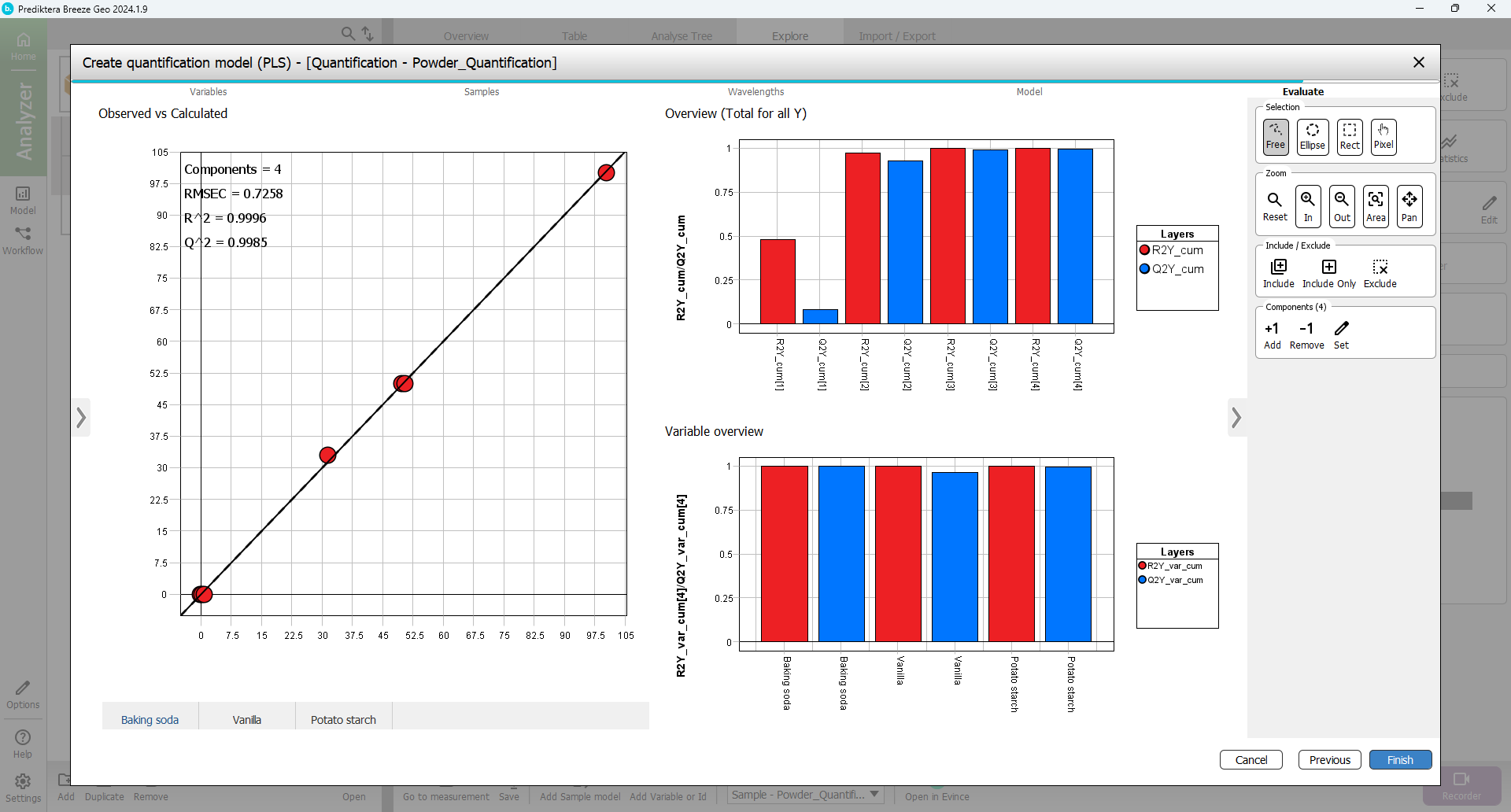
In the last step of the wizard, you can evaluate how good the model is. The “Baking soda” vs. “Ycalc.Baking soda” plot is showing how well the model can explain the variation in this variable. Under the plot are tabs to select the Y variable (Baking soda, Vanilla, Potato starch) to display.
The “Variable overview” is showing the R2 and Q2 for each variable. Everything looks OK so Press “Finish” to complete the model.
Create prediction workflows to quantify new samples
In this step, you will use the quantification model to analyze the % of the three powders in images with samples of unknown content. Press the “Workflow” button in the lower right corner to move from the “Model” mode to the “Workflow” mode.

Press the “Add” button to make a new workflow

In the window that appears, select the “Record data” tab and select the “Unknown Mix” Group. Write a name for the workflow or just use the default.
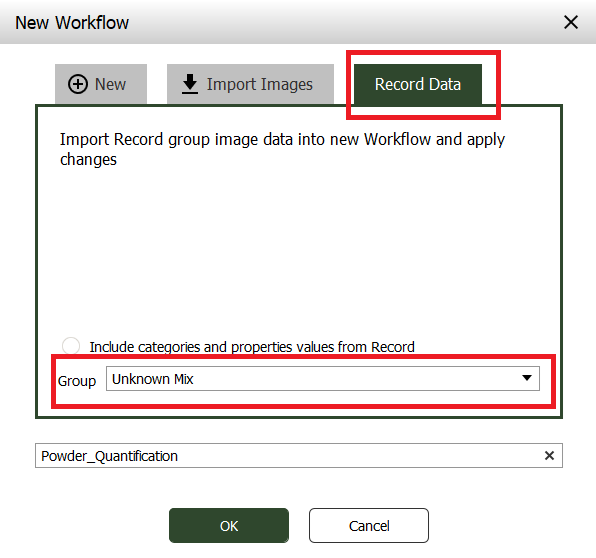
A new workflow will be generated based on the models you have created for this project (sample and quantification models). The images from the “Unknown Mix” group in Record will be imported and applied to this workflow.
Press OK.
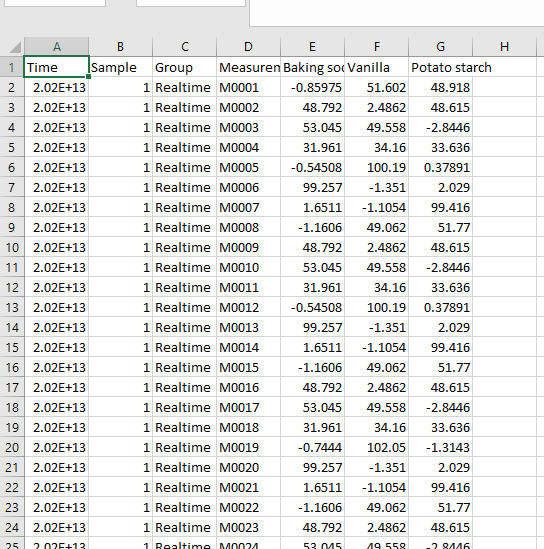
A table is generated with the predicted values (%) for the properties (Baking soda, Vanilla, Potato starch) for the three samples in the “Unknown Mix” images.
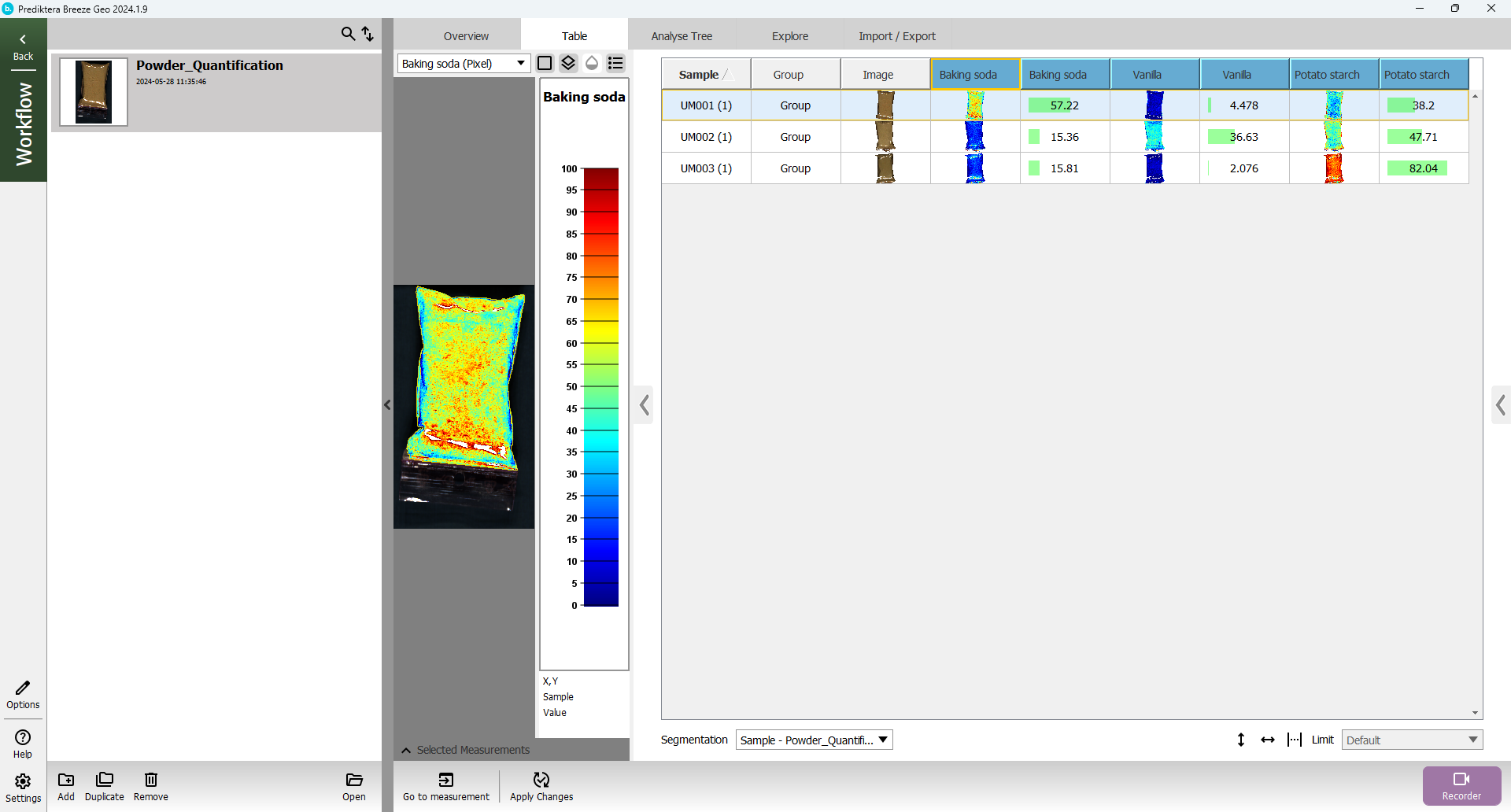
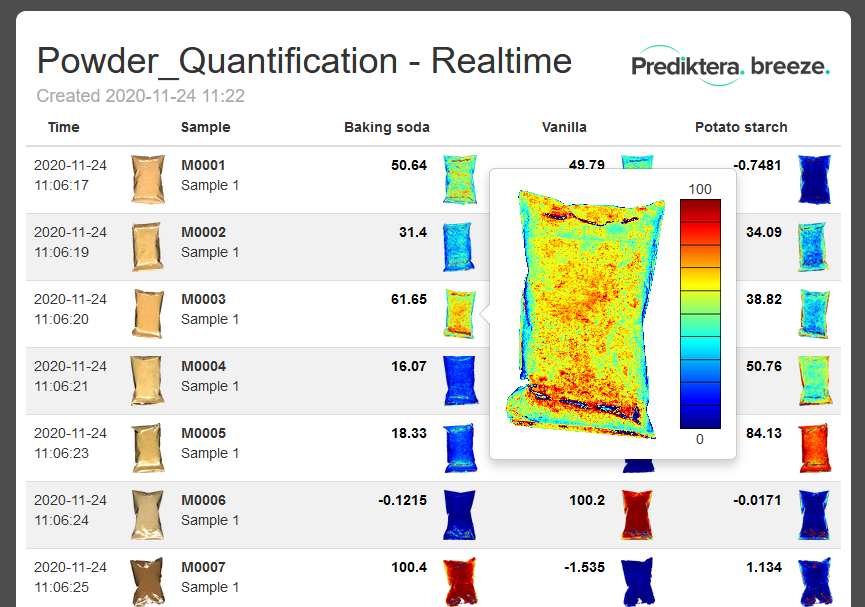
Click on a sample in the “Baking soda” column to color the preview image by the predicted values.
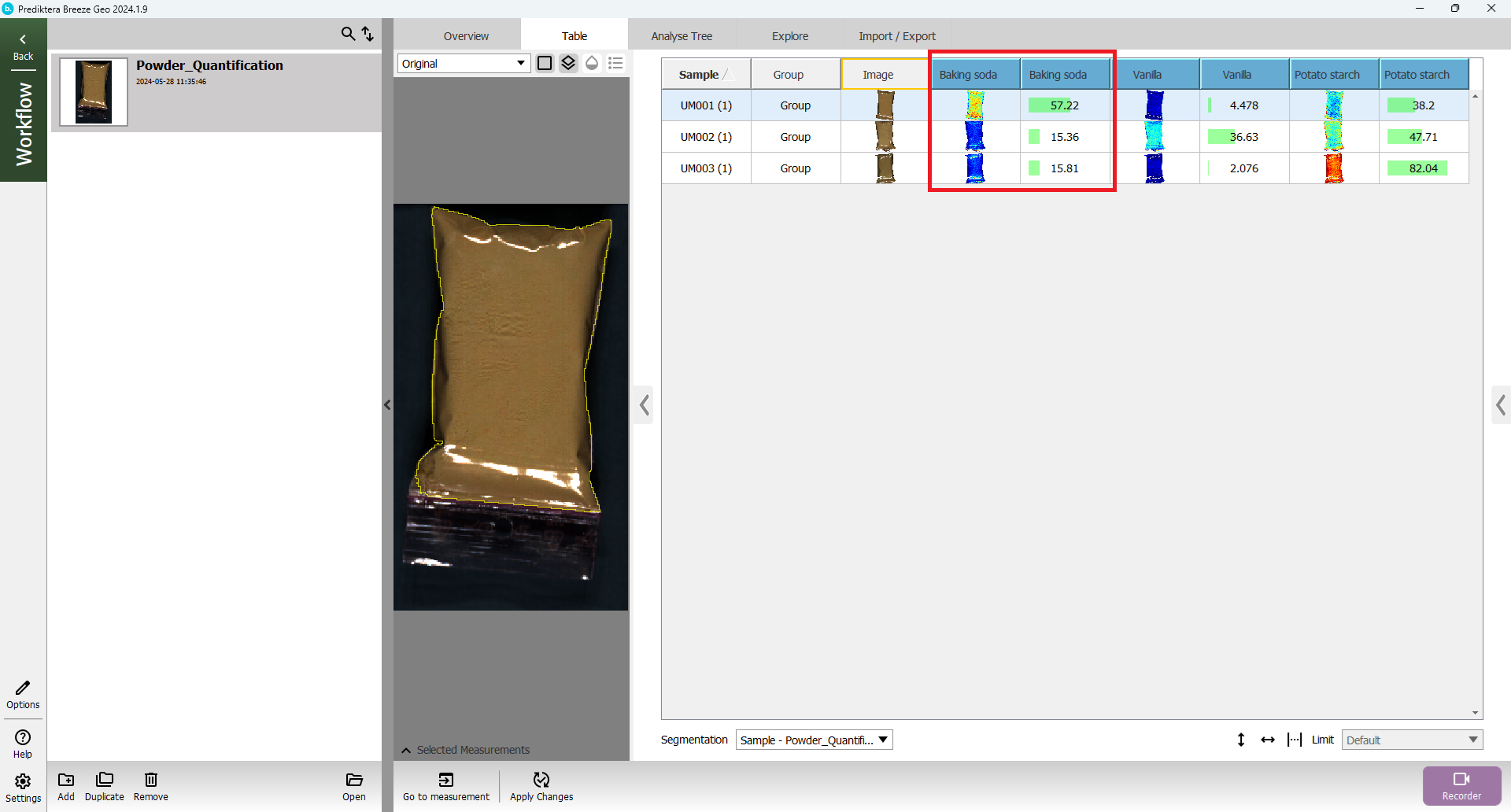
Click on the “legend” button above the preview image to add a legend with the scale.

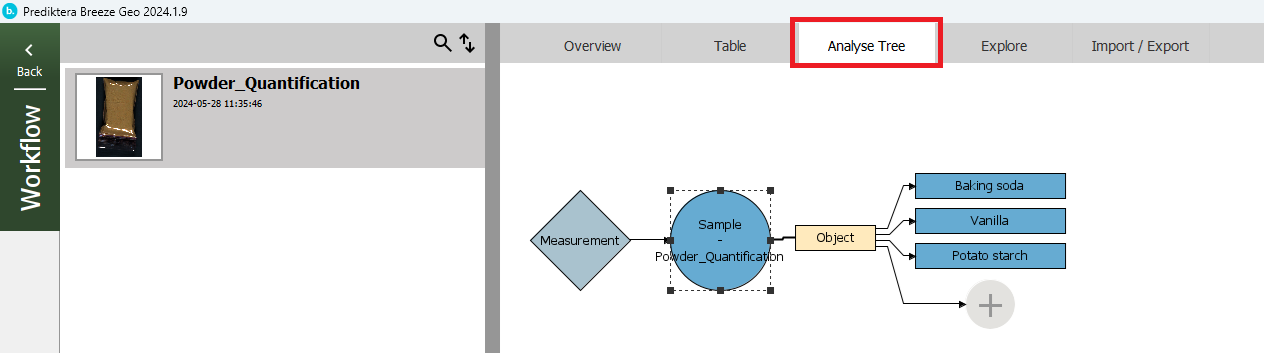
Press the “Analyse Tree” tab to see the steps in the “Workflow”. First, the “Measurement” (image) is analyzed by your sample model (“Sample - Powder Quantification”) to find the sample “Object”. For this object, it then applies your quantification model to calculate the variables (Baking soda, Vanilla, Potato Starch).
Real-time predictions
In addition to analyzing images that are already recorded on your hard drive, you can also use Breeze to analyze images in real-time directly from the camera. If your computer is not connected to a camera, you can simulate this by using the camera simulator in Breeze. With this, it will read images from your hard drive and analyze them continuously. By default, it will use the measurements from the current Record project as input.
In the “Play” mode and with your workflow selected, Press “Analyse”
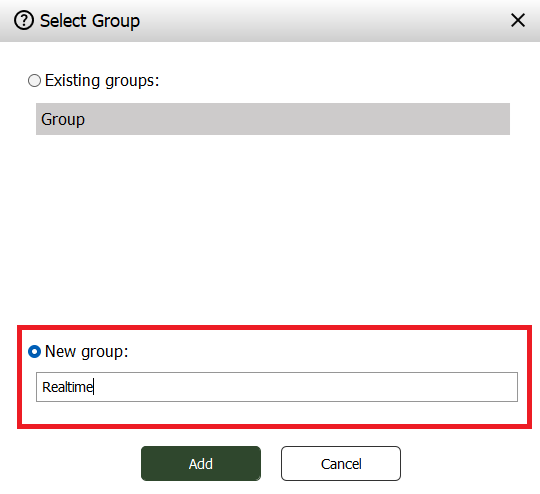
In the window that appears select “New Group”, give it a name like “Realtime” and press “Add”.
In the next step of the Play wizard you can select if you want to save the spectral raw data (spectral data for all pixels) for the images being analysed. If you are scanning many images you can uncheck this option to save storage space. Since these image files are small you can leave this checked and press “Next”.
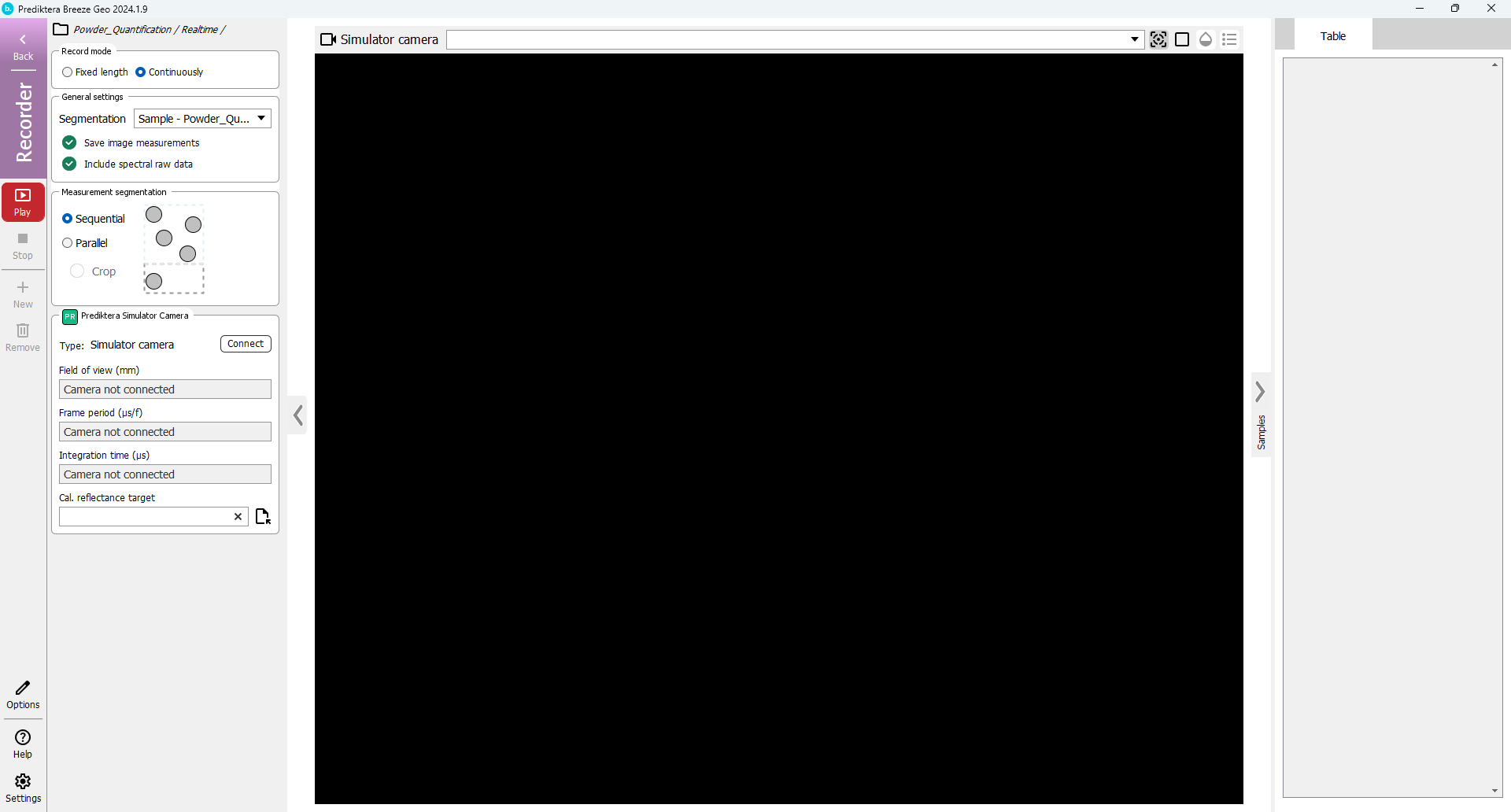
Press “Start”

As you can see the image is analyzed in real-time and the results are displayed in the table. It will loop the 10 images from your Project.
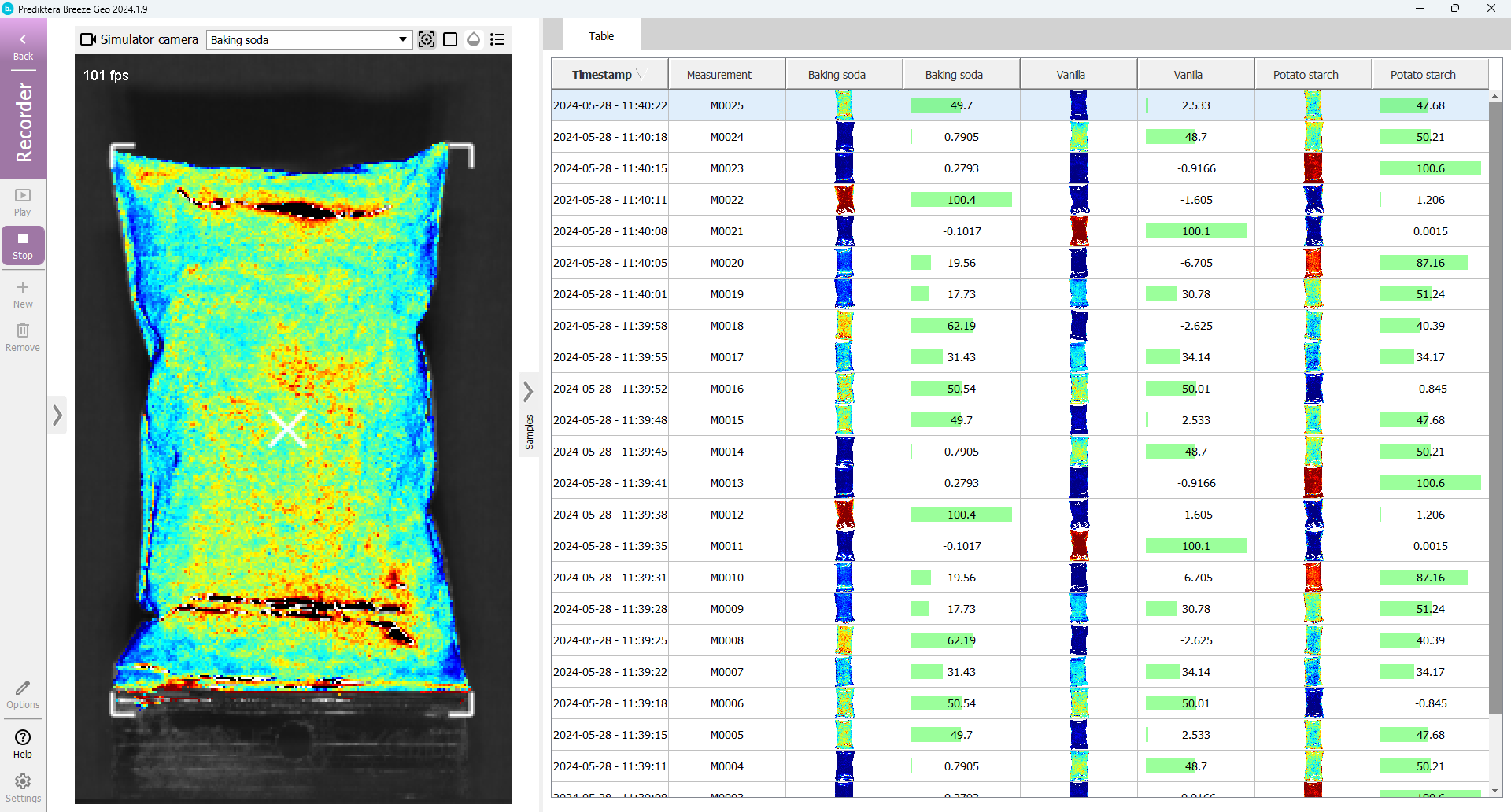
You can change the variable that is displayed by using the drop-down menu above the image, and pressing the button to add a legend.
Press “Stop” to stop the analysis.

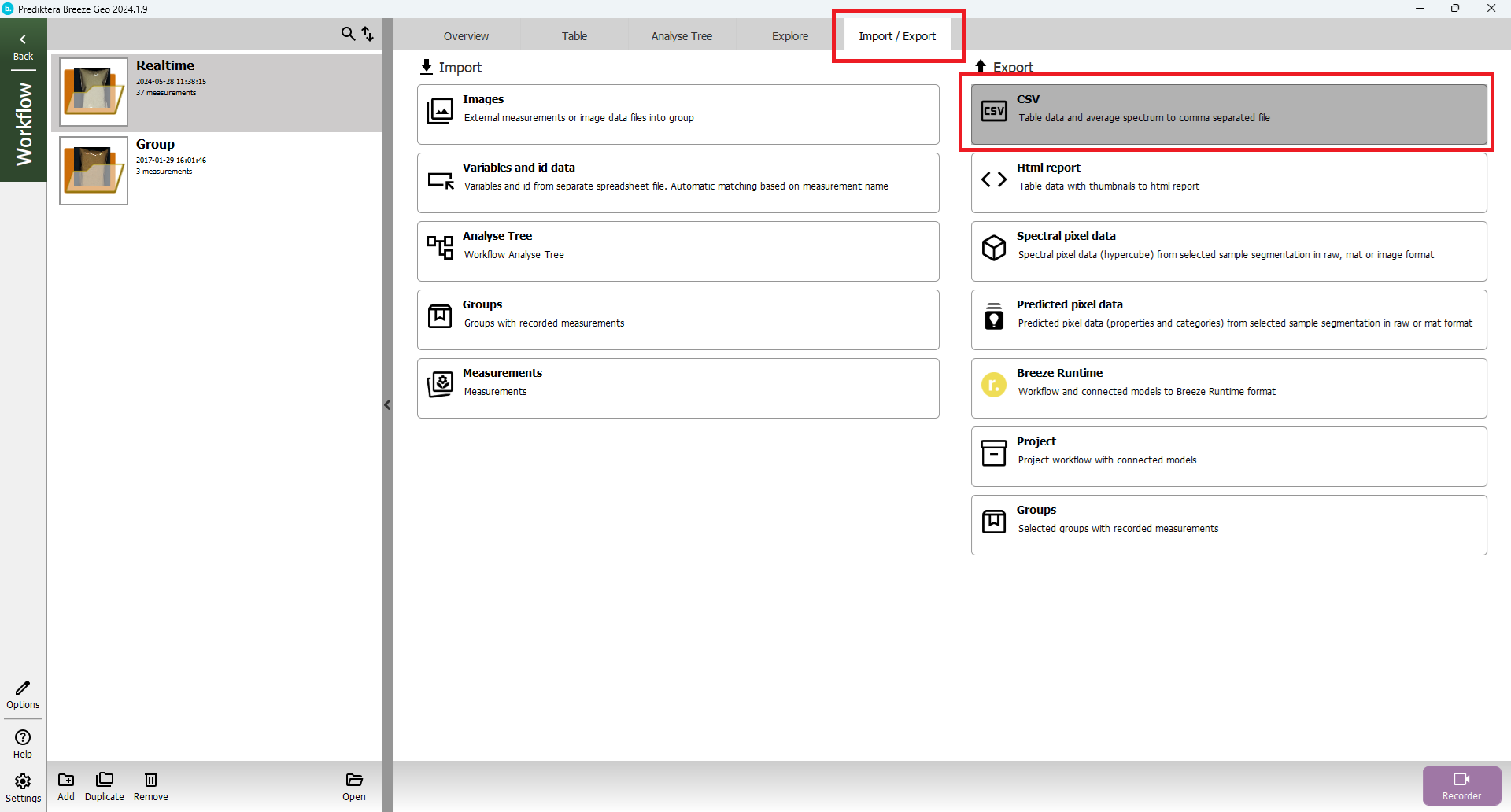
Export to CSV and HTML report
At the group level press the “Export” tab. Here you can for example select to export the results from the “Table” to a .CSV spreadsheet (with the object spectral data) or to an HTML report. You can also export the spectral and predicted pixel data.
Nice job! You have reached the end of step 1 of the “Quantification of Powder” tutorial.
If you would like to learn more please try:
Intro to Breeze: Classification of nuts step 1 or Segmentations and Descriptors
